FIG 3: Mass spectrometry and microscopy methods as tools for detecting and quantifying protein translocations.
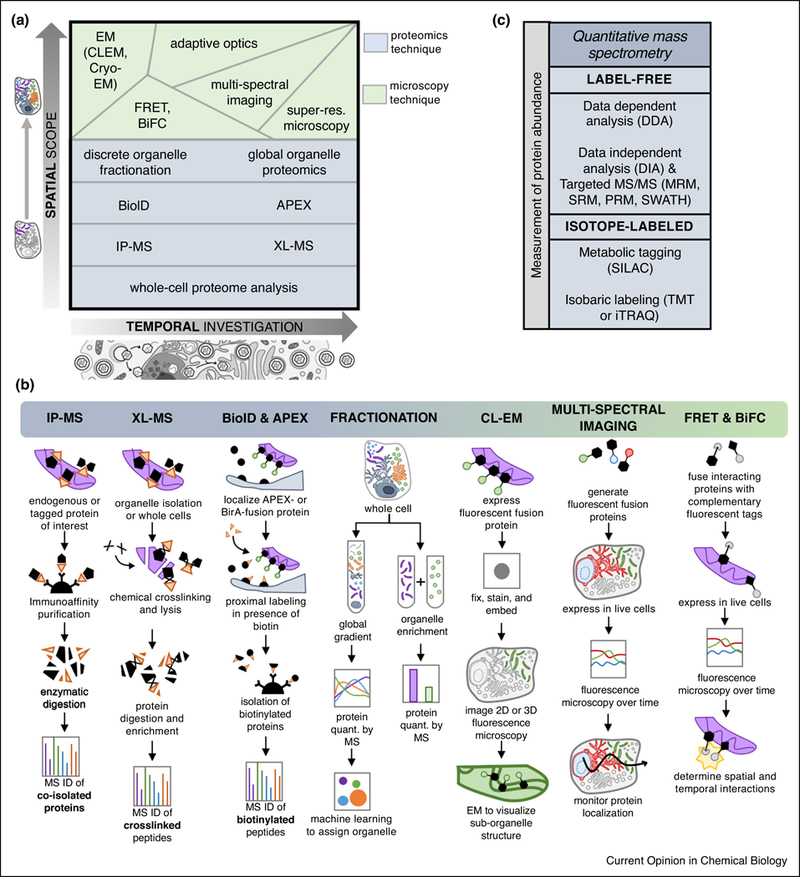
A) Varying levels of spatial scope (i.e. single protein versus single organelle versus global cellular analysis) and temporal capability (i.e. one versus many time points of infection) provided by several mass spectrometry and microscopy methods. B) Schematic representations of selected MS-based proteomic workflows and microscopy methods used in protein translocation studies. C) Quantitative MS approaches can be used to monitor the abundance of proteins in spatial-temporal proteomic studies. Abbreviations: electron microscopy (EM), correlative light EM (CLEM), Förster resonance energy transfer (FRET), bi-molecular fluorescence complementation (BiFC), ascorbate peroxidase (APEX), chemical crosslinking (XL)-MS, immunoaffinity purification (IP)-MS, multiple reaction monitoring (MRM), selected reaction monitoring (SRM), parallel reaction monitoring (PRM), sequential windowed acquisition of all theoretical fragment ion mass spectra (SWATH), stable isotope labeling of amino acids in culture (SILAC), tandem mass tag (TMT), isobaric tags for relative and absolute quantitation (iTRAQ).
