Figure 1. Alternative splicing of hTERT as a mechanism of telomerase regulation in long-lived larger mammals.
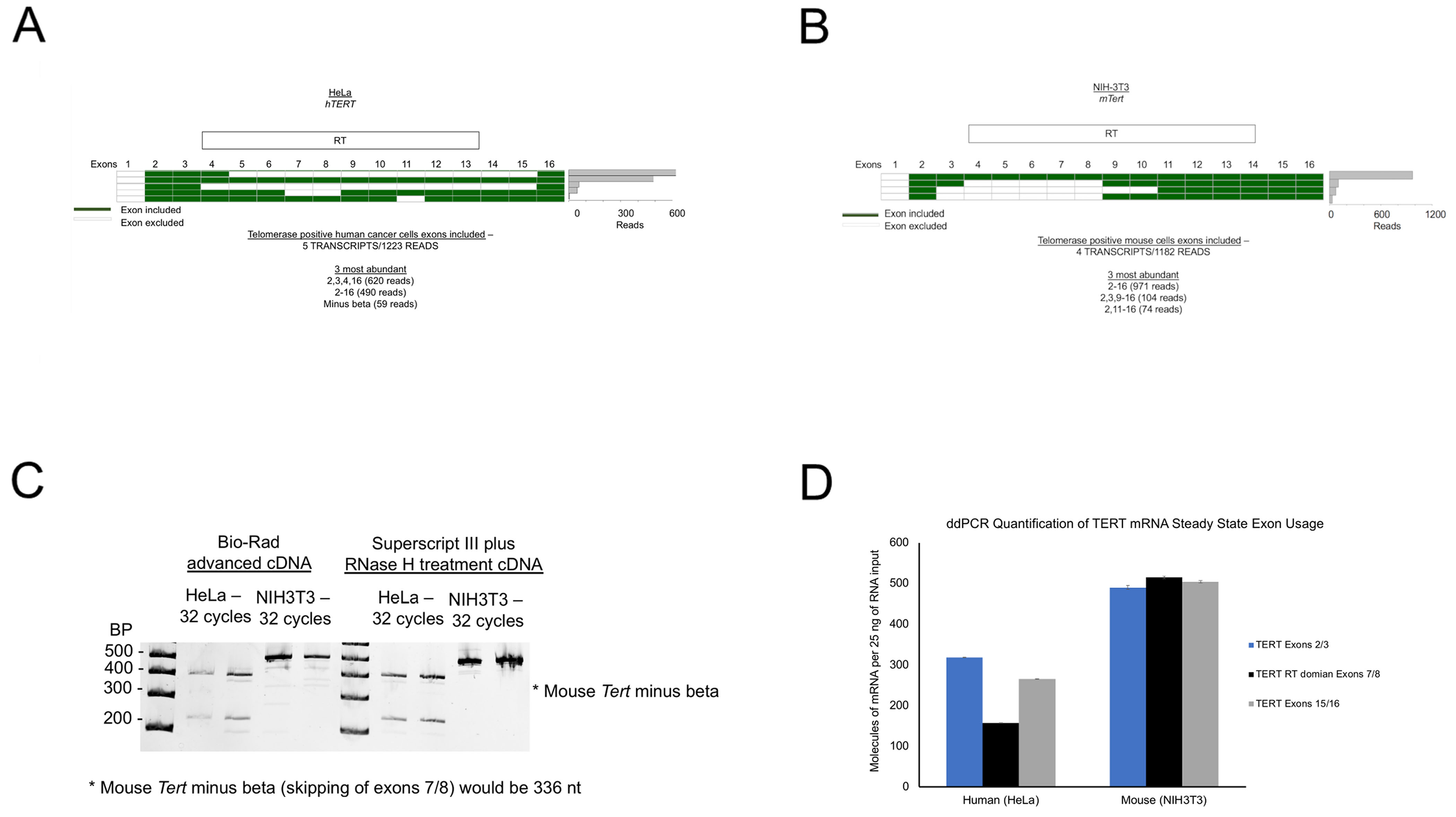
Long-read sequencing of TERT specific libraries was performed to analyze the splicing isoforms of TERT between humans and mice. (A) Human TERT libraries were generated with primers in exon 2 and exon 16 and sequenced by long-read sequencing (Pacific Biosciences). A heatmap of included versus excluded exons is shown. A frequency plot displays the number of reads each isoform that was observed. (B) Mouse Tert libraries were generated with primers in exon 2 and exon 16 and sequenced by long-read sequencing (Pacific Biosciences). A heatmap of included versus excluded exons is shown. A frequency plot displays the number of reads each isoform that was observed. (C) Mouse (NIH3T3) and human (HeLa) cDNAs were amplified with PCR primers spanning TERT exons 5 through 9 of the TERT reverse transcriptase domain (species specific primers were used). N = 3 per species, representative image shown. (D) Species specific TERT quantification across the gene body at three different regions of TERT using droplet digital RT-PCR. N = at least 6 replicates per primer pair.
