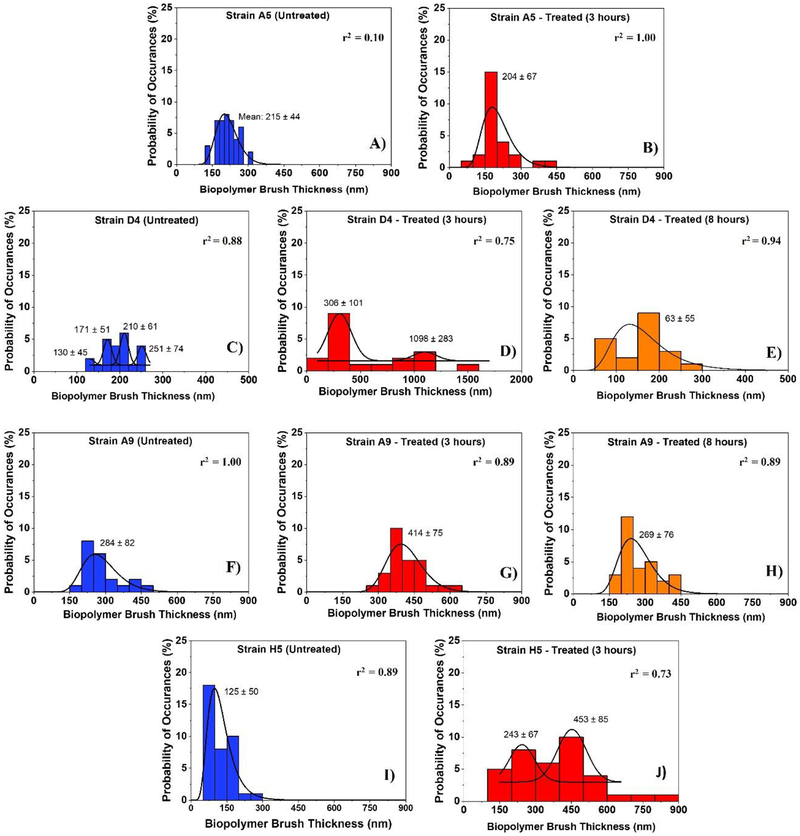
Figure 3:
Histograms showing the distributions of biopolymer brush thicknesses of the MDR-E. coli strains estimated using the steric model for untreated cells (negative control) and for treated ones at different time points (3 and 8 hours). Solid lines show the dynamic peak function model fits the data presented in the histograms (Origin v9.4, OriginLab Corp., Northampton, MA). Lognormal peak function was used for unimodal distributions while the Gaussian peak function was used to fit the multimodal distributions (Arslan et al., 2018). Peaks are reported in nm. When more than one peak is present in the histogram, the reported r2 value represents the average for all fits.