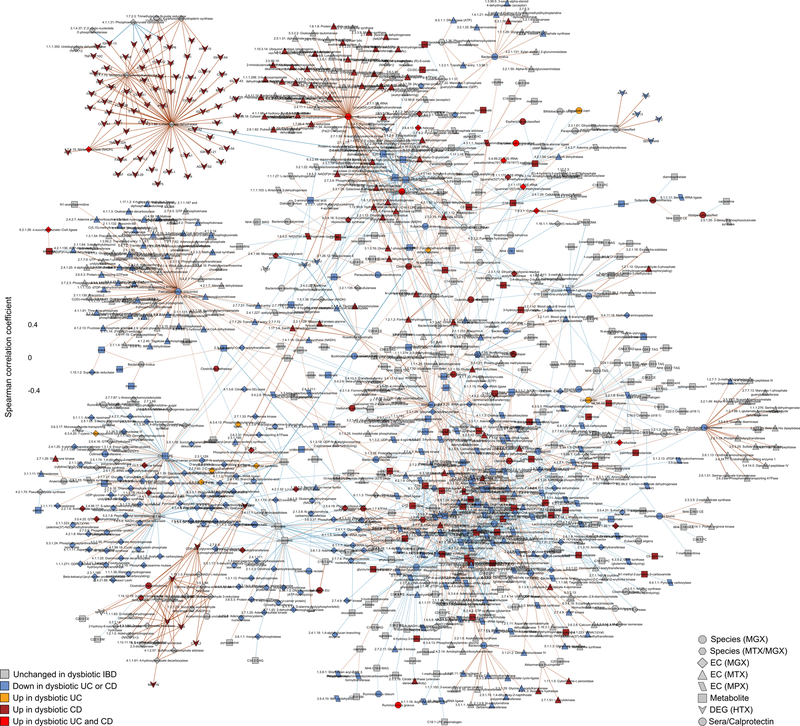
Extended Data Figure 9. Significant covariation among multi’omic components of the gut microbiome and host interactors in IBD (unadjusted).
The network was constructed from 10 datasets: metagenomic species, species-level transcription ratios, functional profiles at the EC levels (MGX, MTX and MPX), metabolites, host transcription (rectal and ileal separately), serology and fecal calprotectin. As in Fig. 4C, measurement types were approximately matched in time with a maximum separation between paired samples of 4 weeks. The top 300 significant correlations (FDR p<0.05) among correlations between features which were differentially abundant in dysbiosis were used to construct the network visualized here (for serology, a threshold of FDR p<0.25 was used). Nodes are colored by the disease group they are “high” in, and edges are colored by the sign and strength of the correlation. For this “unadjusted” network, Spearman correlations were calculated using HAllA from the residuals of the same model as in Extended Data Fig. 8, though without adjusting for dysbiosis (Methods). Appropriate normalization and/or transformation for each measurement type was performed independently before the model fitting (Methods). Singleton node-pairs were pruned from the network. Source associations are in Supplementary Table S36, sample counts in Fig. 1B–C.