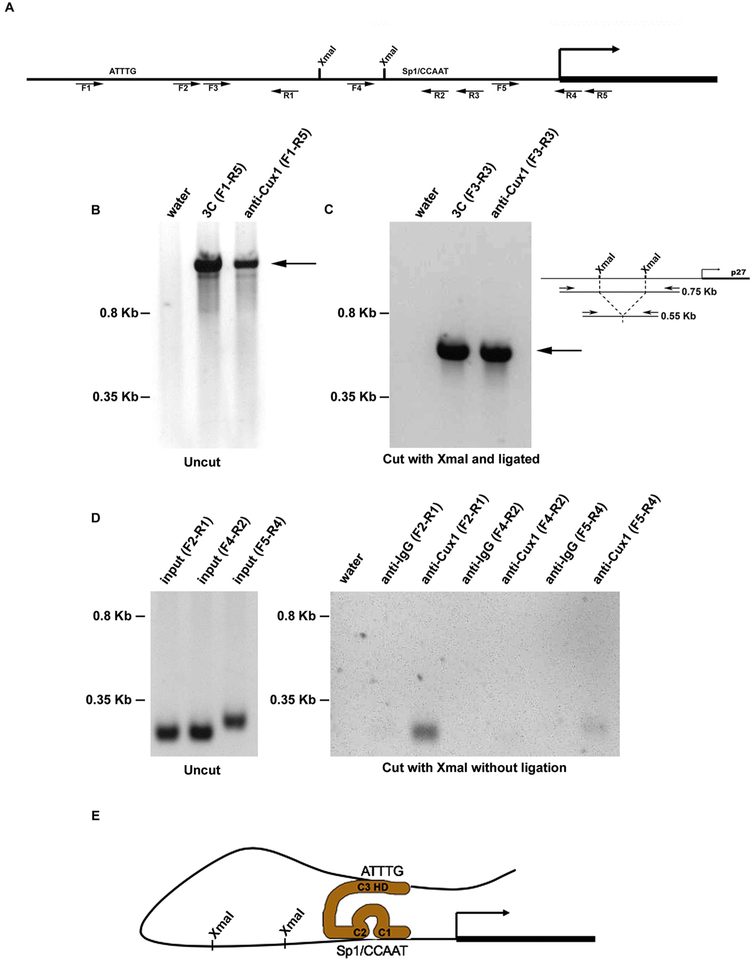
Fig. 1.
Cux1 cross links two sites in the p27 promoter during kidney development. (A) Map of p27 promoter showing the XmaI sites and forward (F) and reverse (R) primers used for 3C and ChIP-Loop experiments used for PCR. The location of sites where Cux1 binds to p27 are indicated by ATTTG and Sp1/CCAAT. Primer sequences are shown in Table 1. (B) Chromatin conformation capture (3C) and ChIP-loop (anti-Cux1) assays were done on genomic DNA containing the p27 promoter region without restriction enzyme digestion. The predicted 1.45 kb fragment of the murine p27 promoter from −1307 to 142 is indicated by the arrow. (C) Chromatin conformation capture (3C) assay amplified a shorter region of DNA looping structure after restriction enzyme digestion and ligation. The 0.55 kb PCR product, indicated by the arrow, is 200 bp shorter than the predicted uncut 0.75 kb DNA, indicating amplification across ligated XmaI sites, shown in the adjacent diagram. Immunoprecipitation with Cux1 antibody (anti-Cux1) after restriction enzyme digestion with XmaI and ligation, followed by reverse cross-linking and amplification, showed the identical 0.55 kb product (arrow), indicating that Cux1 binds the two sites in the p27 promoter simultaneously. (D) Chromatin immunoprecipitation (ChIP) analysis of genomic DNA containing the p27 promoter region following restriction digestion with XmaI shows amplification of a 224 bp fragment of the p27 promoter spanning −940 to −716 (F2-R1) and a 272 bp fragment of the p27 promoter spanning −204 to +68 (F5-R4) relative to the transcription start site and flanking the XmaI sites. In contrast, amplification of a 221 bp fragment of the p27 promoter spanning −483 to −262 (F4-R2), which includes an XmaI site at −383, was observed in the input DNA, but not after immunoprecipitation of XmaI digested DNA with Cux1 antibody. (E) A model of the interaction between Cux1 and the p27 promoter. Cut repeats 1 and 2 compete for CCAAT or Sp1 binding site occupancy, preventing binding by the corresponding transcriptional activators, while the homeodomain binds to AT rich regions. Adapted from (Nepveu, 2001).