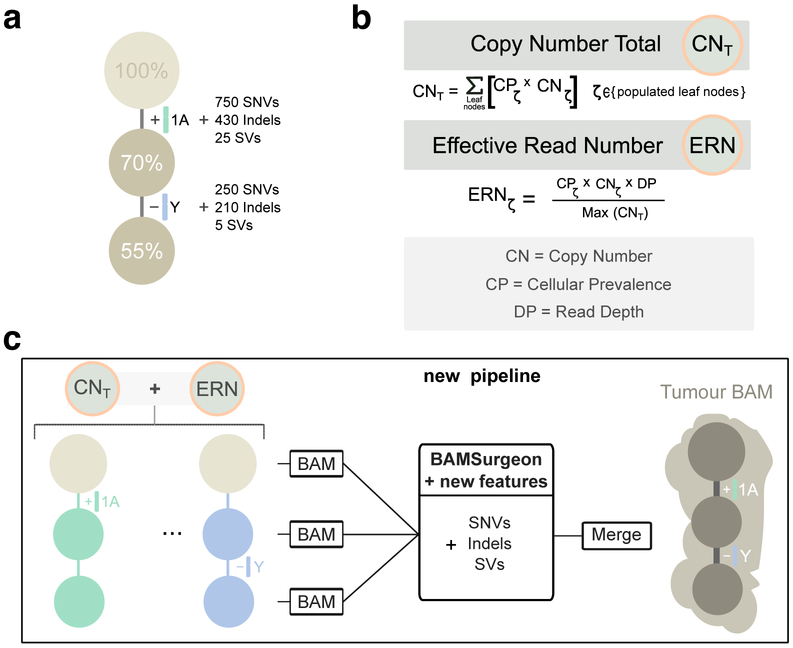
Figure 3 ∣. Simulating subclonal CNAs in tumor BAM files and spiking somatic mutations.
Example case of read number adjustment to simulate subclonal copy number aberrations (CNAs). (a) Desired structure of the tumour being simulated. (b) Read number adjustment calculations. The copy number total (CNT) for each chromosome is its copy number by adjusted by node cellular prevalence summed across all nodes. The maximum CNT across the genome is retained to normalise copy number for all chromosomes. The number of reads assigned to each chromosome at each node (the chromosome’s effective read number) is then computed as the product of the node’s cellular prevalence, the chromosome’s copy number, and the total tumour depth normalised by the maximum CNT. (c) Separation per chromosome phase and per node and new pipeline to simulate tumour BAM files with underlying intra tumour heterogeneity. The first tumour clone (70% CP) has a gain in one copy (referred to as copy A) of chromosome 1 and one of its descendant subclones (55% CP) bears a loss of the Y chromosome. After adjusting read number for CNAs in each BAM corresponding to a node, BAMSurgeon spikes in additional mutations including the new features (complex structural variants, SNVs with trinucleotide contexts and replication timing effects, etc.), and then merges the extracted reads into a final tumor BAM file.