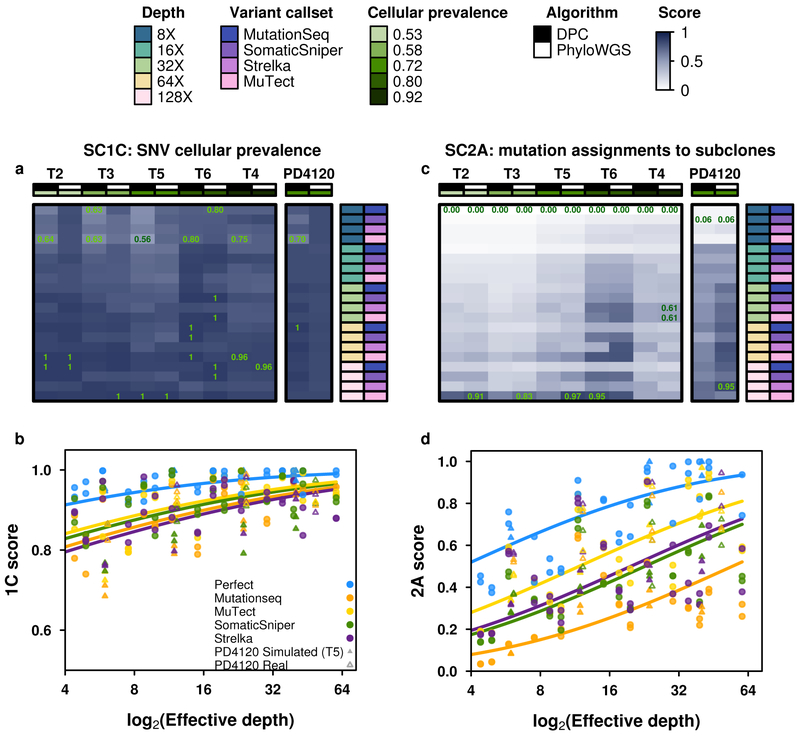
Figure 5 ∣. Error profiles of subclonal reconstruction algorithms.
To identify general features of subclonal reconstruction algorithms, we created a set of tumour-depth-CNA-SNV-subclonal reconstruction algorithm combinations by using the framework outlined in Figure 3 and 4 to simulate five tumours with known subclonal architecture, followed by evaluation of two CNA detection approaches, five SNV detection methods, five read-depths and two subclonal reconstruction methods. The resulting reconstructions were scored using the scoring harness described in Figure 2, creating a dataset to explore general features of subclonal reconstruction methods. All scores are normalised to the score of the best performing algorithm when using perfect calls at the full tumour depth. Scores exceeding this baseline likely represent noise or overfitting and were capped at 1. Only scores from reconstructions using down-sampled CNAs are shown (n=300 tumour-SNV-depth-subclonal reconstruction algorithm combinations). (a) For SC1C (identification of the number of subclones and their cellular prevalence), all combinations of methods perform well. (b) By contrast, for SC2a (detection of the mutational characteristics of individual subclones), there is large inter-tumour variability in performance. (c) Score for SC1c (same as a) as a function of effective read-depth (depth after adjusting for purity and ploidy) improves with increased read-depth, and also changes with the somatic SNV detection method, with MuTect performing best, but still lagging perfect SNV calls by a significant margin. (d) Scores in SC2A show significant changes in performance as a function of effective read-depth.