FIGURE 7. Landscape of Parkin-dependent ubiquitylation and USP30-dependent deubiquitylation on mitochondria in iNeurons.
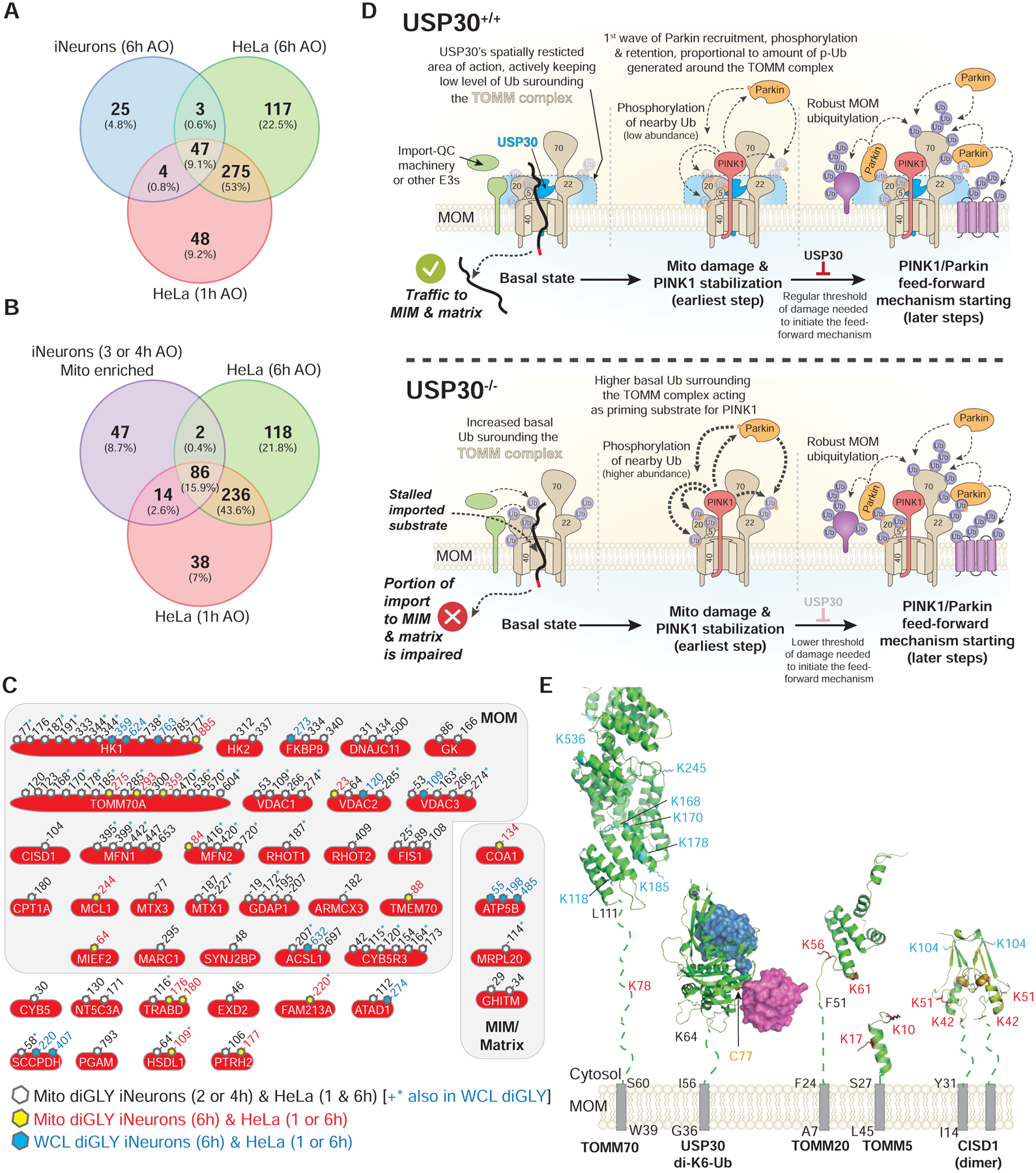
(A) Venn diagram of overlapping diGLY sites observed in whole cell lysates (WCL) (6h post-depolarization) from iNeurons and sites observed for purified mitochondria for HeLa cells 1 or 6h post depolarization. All peptides employed were increased by at least 1-fold (Log2-Ratio>1.0) with p-value <0.05.
(B) Venn diagram of overlapping diGLY sites observed in purified mitochondria (3 and/or 4h post-depolarization) from iNeurons and sites observed for purified mitochondria for HeLa cells 1 or 6h post depolarization. All peptides employed were increased by at least 1-fold (Log2-Ratio >1.0) with p-value <0.05.
(C) Diagram showing the sites of ubiquitination in 41 proteins identified as high confidence Parkin substrates in iNeurons and the corresponding identifications in previously reported data using HeLa cells ectopically expressing Parkin with a parallel TMT-MS3 platform (Rose et al., 2016). Residue numbers for diGLY modified Lys residues are shown. Black font and open hexagon – diGLY site found in purified mitochondria from iNeurons (3 and/or 4h post-depolarization, Table S2) and in purified mitochondria from HeLa cells (1 or 6h post-depolarization). Sites in black font also noted by the blue asterisk were also found in whole cell lysates from iNeurons (6h post-depolarization, Table S1). Red font and yellow hexagon – diGLY site found in purified mitochondria from iNeurons (3 and/or 4h post-depolarization, Table S2) and in purified mitochondria from HeLa cells depolarized for 1h or 6h. Blue font and blue hexagon – diGLY site found in whole cell lysates from iNeurons (6h post-depolarization, Table S1) and in purified mitochondria from HeLa cells depolarized for 1h or 6h.
(D) Model for USP30-dependent mitochondrial protein deubiquitylation.
(E) Structural constraints for USP30-dependent removal of Ub from MOM proteins. Transmembrane segments, grey rectangles. Membrane proximal linkers, dotted lines. Sites of ubiquitylation (red) are those that are reversed by USP30 under depolarization conditions, while those in cyan are largely unchanged in USP30−/− cells. USP30 catalytic cysteine, yellow. PDB codes: TOMM70, modelled on S.c. Tom70, 2GW1; USP30 in complex with K6-di-Ub, 5OHP; CISD1, 3EW0; TOMM5 model, 3PRM; TOMM20 model, 1OM2.
