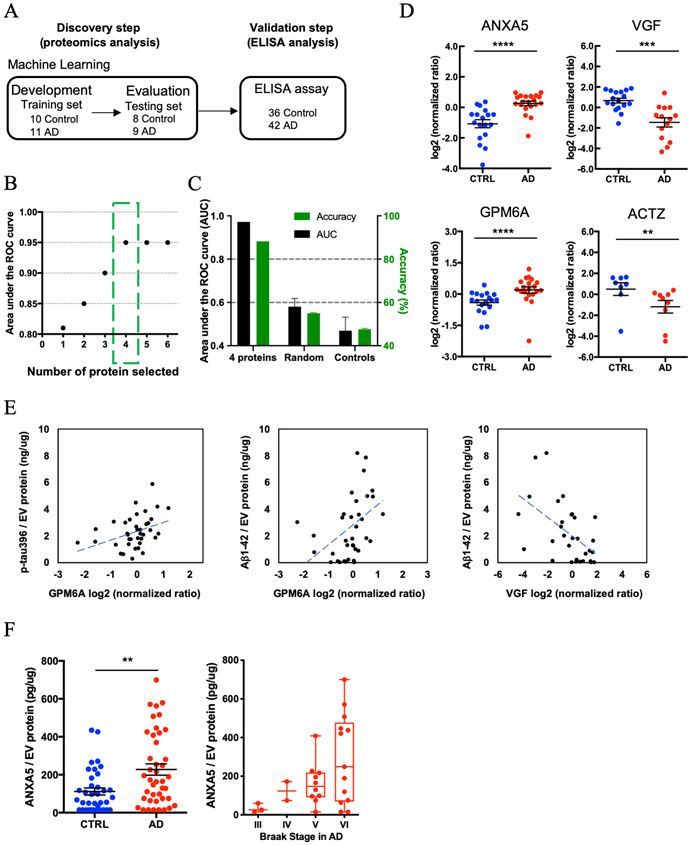
Figure 4. Machine learning model to identify AD brain-specific EV biomarker molecules:
A) Workflow for Machine learning approach. The training set is fed into linear discriminant analysis (LDA) to generate LDA vectors, which are applied to the blinded test set for classification. The predicted molecule are validated from the validation cohort by a commercial ELISA. B) The best performing panel based on the area under the ROC curve using the Lasso algorithm was selected in the training set. Y-axis; the area under the ROC curve, X-axis; proteins selected by the Lasso algorithm. The green box shows the selected protein with High AUC for the blinded test set. C) The Accuracy for 4 selected using the test set. Randomly selected control: Accuracy = 55%, AUC = 0.58., Shuffling control: Accuracy = 47.6%, AUC=0.47. D) A scatter plot of log2 (normalized ratio) as measured by proteomics per selected candidate protein in Machine Learning. (ANXA5: p = 4.58E-06, log2 fold change = 1.1, VGF: p = 5.30E-04, log2 fold change = −1.6, GPM6A: p = 5.37E-04, log2 fold change = 0.6, ACTZ: p = 4.44E-03, log2 fold change = −1.5). The t test was caluculated by Mann-Whitney test. E) Scattered plot of candidate molecules and AD pathogenic molecule in brain-derived EVs. Left: GPM6A and pS396 tau (r = 0.380, p = 0.019 using two-tailed t-test). Center: GPM6A and Aβ1-42 (r = 0.387, p = 0.016). Right: VGF and Aβ1-42 (r = −0.538, p = 0.002). F) Left: Significant difference in total ANXA5 to total EV protein between AD and CTRL group by ELISA (p = 0.0042 by Mann-Whitney test) (AD = 42, CTRL = 36). Right: Braak stage-dependent increase in the ANXA5 expression level to total EV protein in AD dependent manner.