Figure 1. Membrane interactions of peripheral membrane proteins and nMS-compatible vehicles.
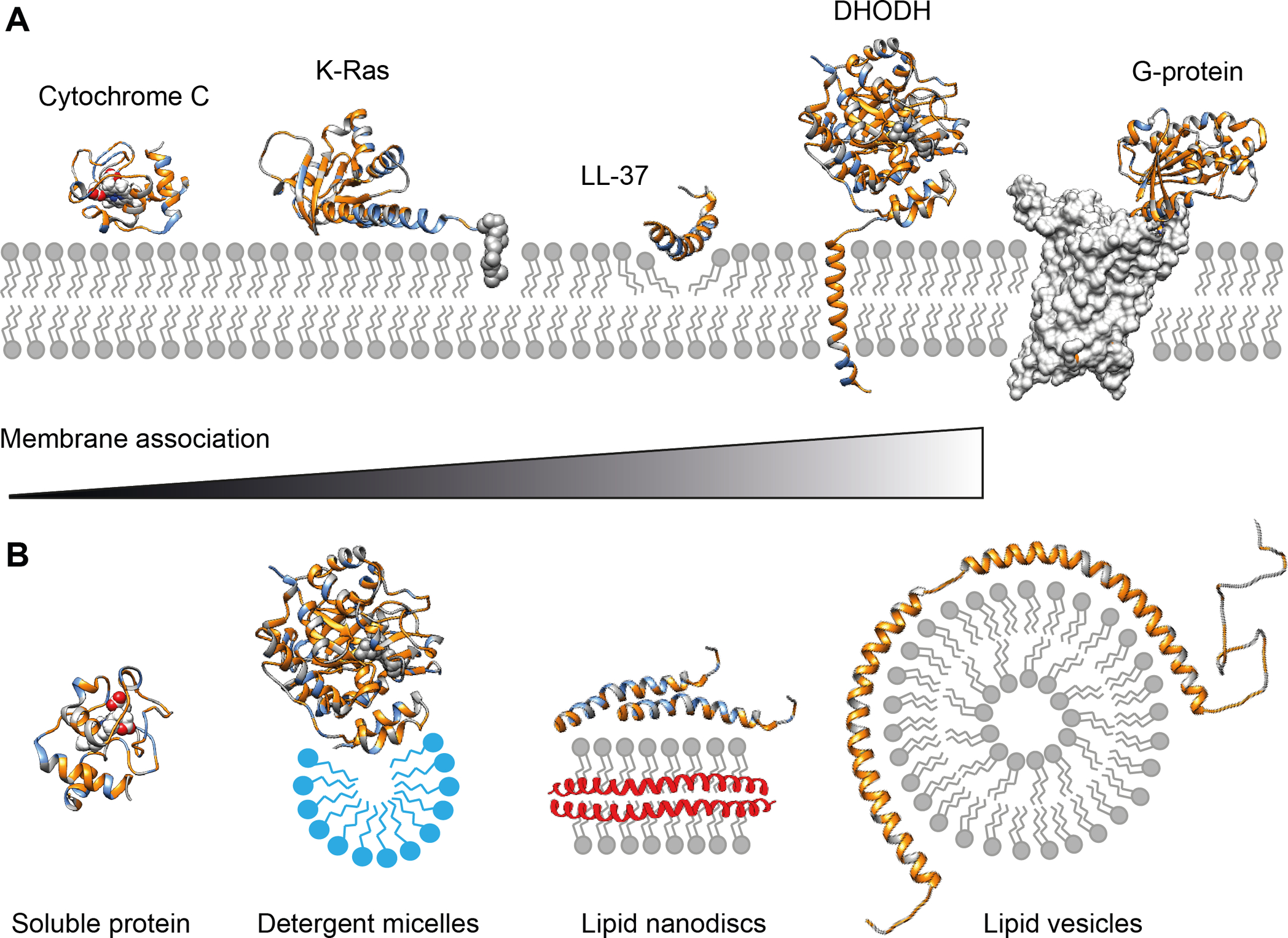
(A) Peripheral membrane protein can associate with the membrane in a variety of ways. Soluble proteins can bind to the membrane via charge and hydrophobic interactions, as exemplified by cytochrome C (PDB ID 1J3S). Additionally, covalent lipid anchors, for example on the C-terminus of K-Ras, can induce a more permanent membrane association (PDB ID 5TAR). The membrane-active peptide LL-37 (PDB ID 2K6O) binds to negatively charged phospholipid head-groups before inserting hydrophobic sidechains into the membrane core to induce pore formation. The mitochondrial enzyme DHODH (PDB ID 2PRM) has an N-terminal transmembrane helix to ensure correct orientation of the substrate binding site towards the membrane. G-proteins (PDB ID 5G53) are not membrane-associated themselves but require individual phospholipids such as PIP2 to form complexes with their membrane-inserted GPCR targets. Hydrophobic regions are rendered in gold, positively charged positions are shown in blue. (B) Strategies to stabilize peripheral membrane proteins for nMS. Although soluble proteins such as cytochrome C can be analyzed by nMS from aqueous solutions, membrane-associated DHODH requires detergent micelles (blue) for stability. Lipid nanodiscs can be used to preserve protein interactions that form on the membrane surface, for example in LL-37 oligomerization. Extensive membrane interactions can be studied using lipid vesicles that provide a large surface for interactions, for example with the amphipathic helices of α-synuclein (PDB ID 2KKW).
