Abstract
Entosis is a type of regulated cell death that promotes cancer cell competition. Though several studies have revealed the molecular mechanisms that govern entosis, the clinical and genetic correlates of entosis in human tumors is less well understood. Here we reviewed entotic cell-in-cell (CIC) patterns in a large single institution sequencing cohort (MSK IMPACT clinical sequencing cohort) of more than 1600 human pancreatic ductal adenocarcinoma (PDAC) samples to identify the genetic and clinical correlates of this cellular feature. After case selection, 516 conventional PDACs and 21 ASCs entered this study and approximately 45,000 HPFs (median 80 HPFs per sample) were reviewed; 549 entotic-CICs were detected through our cohort. We observed that entotic-CIC occurred more frequently in liver metastasis compared to primary in PDAC. Moreover, poorly differentiated adenocarcinoma or adenosquamous carcinoma had more entotic-CIC than well or moderately differentiated adenocarcinoma. With respect to genetic features TP53 mutations, KRAS amplification and MYC amplification were significantly associated with entosis in PDAC tissues. From a clinical standpoint entotic CICs were independently associated with a poor prognosis by multivariate Cox regression analysis when considering all cases or primary PDACs specifically. These results provide a contextual basis for understanding entosis in PDAC, a highly aggressive cancer for which molecular insights are needed to improve survival.
Introduction
Entosis is a type of regulated cell death that originates from actomyosin-dependent cell-in-cell (CIC) internalization and is executed by lysosomes 1. Entosis was first discovered as a nonapoptotic cell death process in 2007 2 and thought to have an advantage for cancer cell survival by promoting cell competition through direct cell-cell interactions unlike other forms of cell death including ferroptosis that occurs at the level of individual cells 3. Entosis is induced microenvironmentally by glucose starvation 4 and has been associated with TP53 mutations 5. Moreover, activated KRAS and Rac signaling have been implicated in establishment of winner cell status and in promoting cell death 6.
While studies in vitro have revealed molecular mechanisms of entosis, translational studies exploring the clinicopathological and genetic correlates of entosis in patients are limited. The available data point to entosis as a correlate of aggressive behavior. In rectal cancer, head and neck squamous cell carcinoma and in lung cancer, a cell in cell (CIC) pattern has been associated with poor prognosis 5,7,8. In breast cancer, the CIC pattern is more prevalent in high grade tumors 9. In pancreatic ductal adenocarcinoma (PDAC), we have recently shown that mutant TP53 status and/or metabolic related gene expression changes are positively correlated with formation of entotic CIC 10. However, the extent to which entosis correlates with metastatic propensity remains unknown, as does the extent to which genes other than TP53 contribute to this phenotypic change. For this reason, we investigated the CIC pattern in PDAC using a large single institution cohort of both primary and metastasis samples for which targeted sequencing data of more than 400 genes are available.
Materials and Methods
MSK IMPACT Clinical Sequencing Cohort Clinical and Genetic Information
All genetic and clinical information and digital whole slide images were obtained from the MSK IMPACT Clinical Sequencing Cohort database through cBioPortal (version2.2.0) 11. Genetic information available for each case was based on results of a targeted sequencing panel for up to 468 known and predicted cancer genes as previously described 12. Treatment information (chemotherapy or chemoradiation therapy) was obtained from an IRB approved and HIPAA compliant RED Cap Database (version 1.0) available through the Center for Pancreatic Cancer Research.
MSK IMPACT Clinical Sequencing Cohort Case Selection
Summary of case selection was shown in Figure 1a. All genetic and clinical information and digital whole slide images were obtained from the MSK IMPACT Clinical Sequencing Cohort database through cBioPortal (version2.2.0) 11. Of the PDAC cases, 1637 (conventional or tubular) ductal adenocarcinomas (PDAC) and 40 adenosquamous carcinomas (ASCs) for which both genetic information and digital images were available for study. All hematoxylin and eosin (H&E) digital slides were reviewed by two gastrointestinal pathologists (A.H. and C.A.I-D.). Samples with massive necrosis and/or degeneration and/or fibrosis, or small biopsy samples with less than 30 evaluable high-power fields (HPFs) was excluded from this study. In addition, in each sample, tumor regions with degeneration and/or necrosis and/or low tumor content (< 10%) were excluded. Finally, a sample set of 537 pancreatic neoplasms (516 conventional PDACs and 21 ASCs) (Figure 1a) were included this study.
Figure 1. Overview of Study Set and Entotic-CICs.
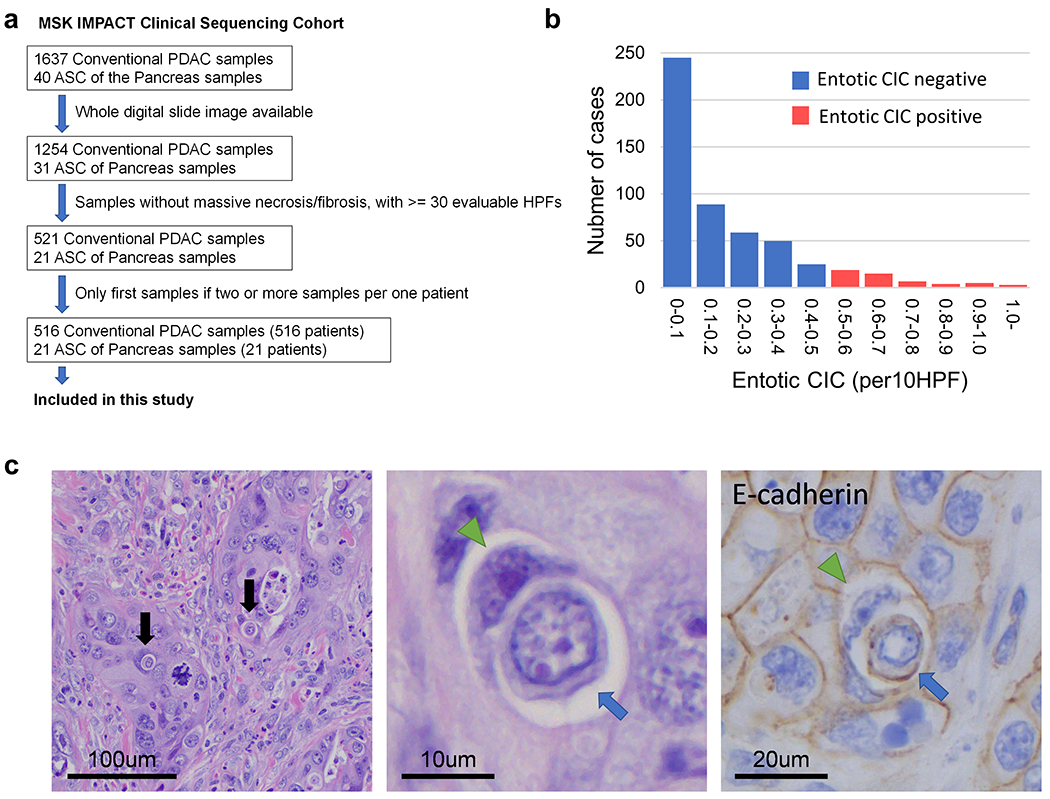
(a) Schematic of sample selection for current study. (b) Distribution of entotic-CIC (per 10 HPFs) in PDACs. (c) Representative histomorphologic and immunohistochemical (E-cadherin) images of entotic-CICs. Entotic-CICs with intervening vacuolar spaces (arrows) were confirmed in low power view. High power view in the middle illustrating a loser cell (blue arrow) engulfed by a winner cell (green arrowhead). E-cadherin confirms entotic-CICs due to the presence of membranous labeling of both the winner and loser cells.
Histologic Definition of Entotic CIC
Entotic cell-in-cell structures (CIC) were defined using H&E images as reported previously 10 as originally proposed by MacKay 5: cytoplasm of the host cell (winner or engulfing cell), nucleus of the host cell (typically crescent-shaped, binucleate, or multilobular and pushed against the cytoplasmic wall), an intervening vacuolar space surrounding the internalized cell (loser), cytoplasm of internalized cell, and nucleus of internalized cell (often round in shape and located centrally or acentrically). If internalized and/or engulfing cells were undergoing typical mitosis or suspected apoptotic changes they were excluded from analysis. Apoptotic changes were characterized by pyknotic nuclei, nuclear fragmentation and loss of nuclear detail.
MSK IMPACT Clinical Sequencing Cohort Slide Review
Digital whole slides available for the 537 cases selected for study from the MSK IMPACT Clinical Sequencing Cohort were visualized through Smart Slide Viewer (version 1.3.1583). All evaluable tumor regions submitted to the IMPACT sequencing in digital slides were reviewed by two pathologists (A.H. and C.I.D.). A total ~45,000 HPFs (range 30–197 HPFs, median 80 HPFs per sample) were reviewed and all potential or suspected entotic-CICs (~1000 unique images) were captured and reviewed with discussion. Entotic-CICs were quantified by calculating the average number per 10 HPFs. Based on this information we divided the cohort into two groups as previously reported 10. “Entotic-CIC positive” PDACs were defined as those with 0.5 or more entotic CICs per 10 HPFs, corresponding to the top ~10% in this study. PDACs not meeting these criteria were defined as “entotic-CIC negative”. Tumor grading (well-, moderately-, poorly differentiated adenocarcinoma) of all cases was determined based on WHO classification 13.
Immunohistochemistry of E-Cadherin
The histologic classification of entosis in 15 representative cases with entotic-CICs from the MSK IMPACT Clinical Sequencing Cohort was validated using immunohistochemistry for E-cadherin 14. Five micro-meter unstained sections were cut from the FFPE blocks of each case and immunolabeled with a mouse monoclonal antibody against E-cadherin (BD Biosciences, Catalog No. 610181, clone 36/E-Cadherin, 5ug/ml) according to optimized protocol on a Ventana Benchmark XT autostainer (Ventana Medical Systems Inc.) in the MSK Pathology Core Lab. Appropriate positive and negative controls were included in each run.
Genetic Features of Cohort
Using cBioportal as a guide the top 10 most frequently mutated genes and the top five most frequent copy number alterations in the cohort of 537 patients were included for correlation to entotic-CICs.
Autopsy Cohort
Matched primary and metastatic tissues from three patients in the Last Wish Program at Memorial Sloan Kettering Cancer Center and 21 patients from the Gastrointestinal Cancer Rapid Medical Donation Program at Johns Hopkins Hospital were used for the validation autopsy cohort. All slides were reviewed by two gastrointestinal pathologists (A.H. and C.I.D.). At least two slides (median 8 slides, range 2-12) and 50 HPFs (median 379 HPFs, range 52-1078) for each primary and metastasis site were evaluated for entotic-CICs. Only metastases from the liver, lung and/or peritoneal cavity were included.
Ethics Statement
This study was approved by the Review Boards of Johns Hopkins School of Medicine and Memorial Sloan Kettering Cancer Center.
Statistics
All statistics and graphs were performed and made using GraphPad Prism (version 8.2.1) and/or XLSTAT (version 2019.4.2). Each analysis method was described in Results or Figure Legends. Statistical significance was considered if P-value is less than 0.05.
Results
Entotic Cell in Cell Structures in PDAC
We identified 539 entotic-CICs within 537 PDAC (516 conventional PDAC and 21 ASC) cases in the MSK IMPACT Clinical Sequencing Cohort. Entotic-CICs per 10 HPFs ranged from 0 to 3.06 (median 0.12) per neoplasm (Figure 1b and 1c). Per our predefined criteria (see Materials and Methods), 70 of 537 carcinomas (13.0%) correspond to entotic-CIC positive PDACs. All (70/70, 100%) entotic-CIC positive cases had two or more entotic-CICs and more than 80% (59/70 cases, 84.3%) of cases had three or more. Representative cell in cell patterns (~50 CICs in 15 cases) were confirmed by E-Cadherin IHC that demonstrated membranous labeling of both the winner cell and the cell being engulfed (Figure 1c).
Clinical Characteristic of Entotic-CIC Positive PDAC
Entotic-CIC positive status was unrelated to age, gender in univariate analysis of the IMPACT cohort (Table 1). No significant differences were seen for entotic CIC positivity between PDAC and ASC in total cohort (Table 1), although when the analysis was limited to primary tumors only poorly differentiated adenocarcinoma (23 out of 111 samples, 20.7%) or ASC (5/19, 26.3%) had more entotic-CIC positivity than well (2/75, 2.7%) or moderately (10/210, 4.8%) differentiated adenocarcinoma tumors (P < 0.0001, two-sided Fisher’s exact test) (Table 2). For primary surgical resected cases, cases with neoadjuvant chemo- or chemoradiation-therapy had less entotic-CIC (3/69, 4.3%) compared to treatment naïve primary tumors (37/340, 10.9%), but this difference was not statistically significant (P = 0.120, two-sided Fisher’s exact test) (Table 2). No differences were confirmed between lymph node positive and negative cases (Table 2). Entotic-CIC positivity was significantly more prevalent in metastases (30 out of 122 samples, 24.6%) compared to unmatched primary tumors (40/415 samples, 9.6%) (P < 0.0001, two-sided Fisher’s exact test) (Table 1). Among metastatic sites, entotic CIC positive samples most often corresponded to liver metastases (24/69, 34.8%) (Table 3).
Table 1.
Clinicopathologic Characteristics of Entotic-CIC Positive PDAC
| Factor | Total | Entotic-CIC |
P-Value | |||
|---|---|---|---|---|---|---|
| Positive | Negative | % Positive | ||||
| Age | > 65 | 317 | 45 | 272 | 14.2% | 0.364 |
| < = 65 | 220 | 25 | 195 | 11.4% | ||
| Gender* | Male | 299 | 39 | 260 | 13.0% | 1.000 |
| Female | 237 | 31 | 206 | 13.1% | ||
| Histology** | PDAC | 516 | 65 | 451 | 12.6% | 0.175 |
| ASC | 21 | 5 | 16 | 23.8% | ||
| Location | Primary | 415 | 40 | 375 | 9.6% | < 0.0001 |
| Metastasis | 122 | 30 | 92 | 24.6% | ||
Gender information was not available for one patient.
PDAC: Pancreatic ductal adenocarcinoma, ASC: Adenosquamous carcinoma
Each P-value was calculated with Fisher’s exact test, two-sided.
Table 2.
Primary Characteristic of Entotic-CIC Positive PDAC
| Factor | Total | Entotic-CIC |
P-Value | |||
|---|---|---|---|---|---|---|
| Positive | Negative | % Positive | ||||
| Tumor Grade | Well | 75 | 2 | 73 | 2.7% | < 0.0001 |
| Moderately | 210 | 10 | 200 | 4.8% | ||
| Poorly | 111 | 23 | 88 | 20.7% | ||
| ASC | 19 | 5 | 14 | 26.3% | ||
| Lymph Node Metastasis* | Positive | 290 | 28 | 262 | 9.7% | 1.000 |
| Negative | 118 | 11 | 107 | 9.3% | ||
| Neoadjuvant Therapy** | Yes | 69 | 3 | 66 | 4.3% | 0.120 |
| No | 340 | 37 | 303 | 10.9% | ||
Lymph node metastasis information is available for 408 surgical resected cases.
Neoadjuvant therapy information is available for 409 surgical resected cases. 48 cases had chemotherapy and 21 cases had chemoradiation therapy.
ASC, adenosquamous carcinoma.
Each P-value was calculated with Fisher’s exact test, two-sided.
Table 3.
Metastatic Characteristic of Entotic-CIC Positive PDAC
| Metastasis Location | Total | Entotic-CIC |
||
|---|---|---|---|---|
| Positive | Negative | % Positive | ||
| Liver | 69 | 24 | 45 | 34.8% |
| Peritoneal Cavity | 25 | 2 | 23 | 8.0% |
| Lung | 12 | 2 | 10 | 16.7% |
| Other* | 16 | 2 | 14 | 12.5% |
Others include metastases of lymph nodes (6), soft tissue (2), brain (1), pleura (1), retroperitoneum (1), arm (1), skin (1), umbilicus (1), unknown (2).
We next reviewed matched primary and liver metastasis samples from an independent cohort of patients who underwent an autopsy. This confirmed that liver metastases had more entotic-CICs than primary carcinomas (P = 0.023, Wilcoxon matched-pair signed rank test, two-sided) (Figure 2a, b). To determine the extent that different sites of metastasis in the same patient exhibit entotic-CICs we further assessed available lung or peritoneal metastases in these patients. This revealed heterogeneity across the primary tumor and metastases in that entotic-CICs in one site were not indicative of a high number of entotic-CICs in another metastatic site in the same patient, though the degree of heterogeneity varied among cases (Figure 2a, c).
Figure 2. Heterogeneity of Entotic-CICs in PDAC.
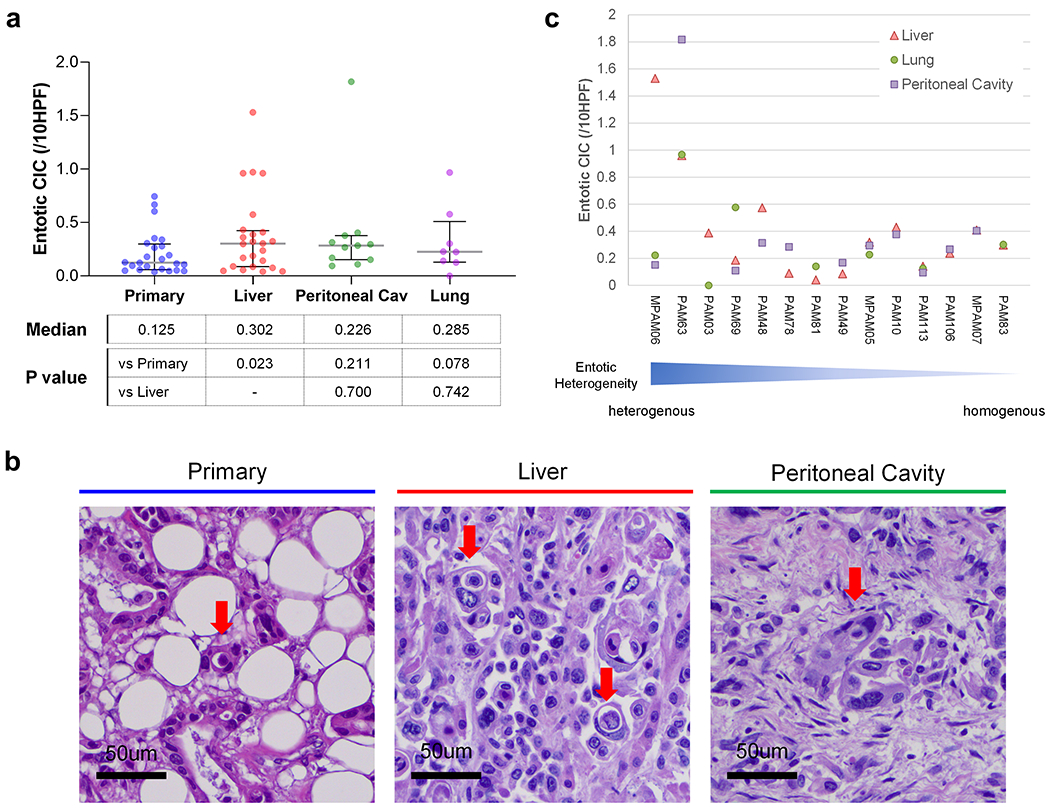
(a) Entotic-CIC (per 10 HPFs) in matched primary, and liver, peritoneal cavity and lung metastasis in end-stage PDAC autopsy cohort. (b) Representative histomorphologic images of entotic-CICs in primary and live and peritoneal cavity metastases in autopsy series MPAM06. More than one entotic-CICs were found in one HPF in liver metastasis. (c) Heterogeneity of entotic-CIC among metastatic organs in end-stage PDAC autopsy cohort.
Genetic Characteristics of Entotic-CIC Positive PDAC
Entotic-CIC positive PDACs had a higher prevalence of TP53 mutations (P = 0.041, Fisher’s exact test, two-sided) and KRAS and MYC amplification (P = 0.025 and 0.012, Fisher’s exact test, two-sided) (Table 4). Deep deletions for CDKN2A, TP53 or SMAD4 were not statistically associated with entotic-CIC positivity (Table 4). To determine if a specific type of TP53 alteration was associated with the formation of entotic-CICs we compared lollipop plots of the mutational spectrum of TP53 in entotic-CIC positive versus negative cases (Figure 3). We found no difference between the prevalence of hot spot mutations (P = 0.331, Fisher’s exact test, two-sided) nor did we find a difference in the proportion of truncating and non-truncating TP53 mutations between the two groups (P = 1.000, Fisher’s exact test, two-sided). (Supplementary Information 1). The distribution of TP53 mutations also did not differ among CIC positive and negative carcinomas among primary and metastatic sites specifically (Supplementary Information 2). While a prior report suggested expression of KRAS-G12V in eating cells is associated with entosis 6, we observed no relationship of a specific KRAS hotspot mutation in our cohort (P = 0.501, Fisher’s exact test, two-sided) (Supplementary Information 3) .
Table 4.
Genetic Characteristics of Entotic-CIC Positive PDAC
| Category | Gene | Total | Entotic-CIC |
P-Value | |||
|---|---|---|---|---|---|---|---|
| Positive | Negative | % Positive | |||||
| Mutation | KRAS | Mutated | 492 | 68 | 424 | 13.8% | 0.102 |
| WT | 45 | 2 | 43 | 4.4% | |||
| TP53 | Mutated | 399 | 59 | 340 | 14.8% | 0.041 | |
| WT | 138 | 11 | 127 | 8.0% | |||
| CDKN2A | Mutated | 137 | 24 | 113 | 17.5% | 0.078 | |
| WT | 400 | 46 | 354 | 11.5% | |||
| SMAD4 | Mutated | 113 | 20 | 93 | 17.7% | 0.115 | |
| WT | 424 | 50 | 374 | 11.8% | |||
| CDKN2AP16INK4A | Mutated | 97 | 15 | 82 | 15.5% | 0.410 | |
| WT | 440 | 55 | 385 | 12.5% | |||
| ARID1A | Mutated | 56 | 10 | 46 | 17.9% | 0.292 | |
| WT | 481 | 60 | 421 | 12.5% | |||
| CDKN2AP14ARF | Mutated | 59 | 10 | 49 | 16.9% | 0.313 | |
| WT | 478 | 60 | 418 | 12.6% | |||
| RNF43 | Mutated | 29 | 4 | 25 | 13.8% | 0.782 | |
| WT | 508 | 66 | 442 | 13.0% | |||
| KDM6A | Mutated | 28 | 5 | 23 | 17.9% | 0.393 | |
| WT | 509 | 65 | 444 | 12.8% | |||
| APC | Mutated | 25 | 2 | 23 | 8.0% | 0.759 | |
| WT | 512 | 68 | 444 | 13.3% | |||
| CNV | MYC | Amplified | 14 | 5 | 9 | 35.7% | 0.025 |
| No amplified | 523 | 65 | 458 | 12.4% | |||
| AKT2 | Amplified | 18 | 6 | 12 | 33.3% | 0.205 | |
| No amplified | 519 | 64 | 455 | 12.3% | |||
| CCNE1 | Amplified | 10 | 3 | 7 | 30.0% | 0.130 | |
| No amplified | 527 | 67 | 460 | 12.7% | |||
| KRAS | Amplified | 8 | 4 | 4 | 50.0% | 0.012 | |
| No amplified | 529 | 66 | 463 | 12.5% | |||
| FGFR1 | Amplified | 6 | 1 | 5 | 16.7% | 0.569 | |
| No amplified | 531 | 69 | 462 | 13.0% | |||
| CDKN2A | Deep deletion | 45 | 6 | 39 | 13.3% | 1.000 | |
| No deletion | 492 | 64 | 428 | 13.0% | |||
| CDKN2AP16INK4A | Deep deletion | 44 | 6 | 38 | 13.6% | 0.818 | |
| No deletion | 493 | 64 | 429 | 13.0% | |||
| CDKN2AP14ARF | Deep deletion | 42 | 6 | 36 | 14.3% | 0.811 | |
| No deletion | 495 | 64 | 431 | 12.9% | |||
| CDKN2B | Deep deletion | 40 | 4 | 36 | 10.0% | 0.806 | |
| No deletion | 497 | 66 | 431 | 13.3% | |||
| SMAD4 | Deep deletion | 18 | 2 | 16 | 11.1% | 1.000 | |
| No deletion | 519 | 68 | 451 | 13.1% | |||
Each P-value was calculated with Fisher’s exact test, two-sided.
Figure 3. Lolliplot of TP53 in Entotic-CIC Positive and Negative PDACs.
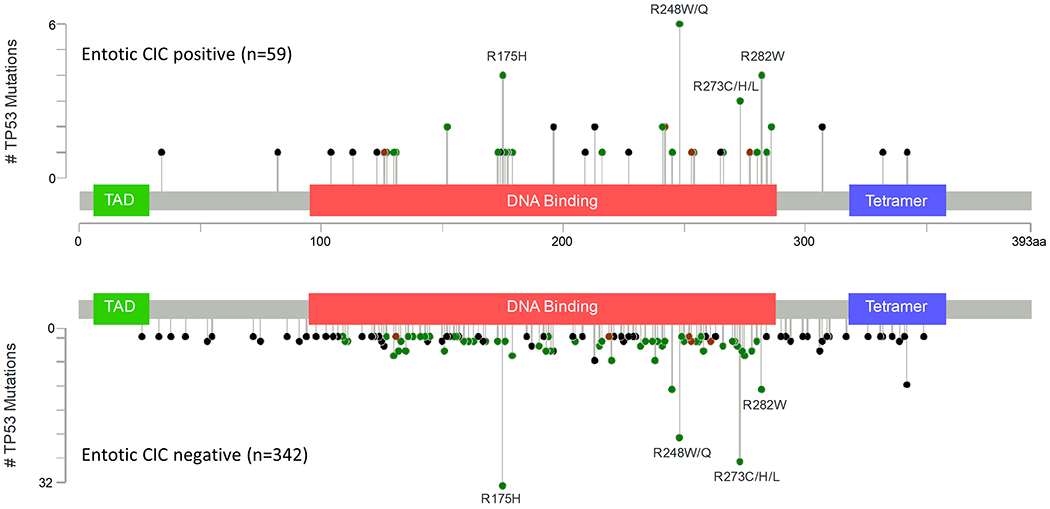
Number of total cases is shown in parentheses.
Outcome of Entotic-CIC Positive PDAC
Kaplan–Meier analysis revealed significant poorer prognosis of entotic-CIC Positive PDAC when considering all cases (P = 0.0002, log-rank test) (Figure 4a) or primary carcinomas only (P = 0.021, log-rank test) (Figure 4b). Cox proportional hazards analysis showed positive entotic-CICs, gender, tumor location and KRAS amplification were all associated with a poor prognosis by both univariate and multivariate analyses (Table 5 and Table 6). Entotic-CIC positive status was an independent prognostic variable by both univariate (P < 0.001, Hazard ratio: 1.813) (Table 5) and multivariate analysis (P = 0.014, Hazard ratio: 1.527) (Table 6). This significance identified by multivariate analysis was also confirmed when we use primary carcinomas only (P = 0.018, Hazard ratio: 1.668) (Table 7).
Figure 4. Kaplan-Meier Analysis of Entotic-CIC Positive and Negative PDAC.
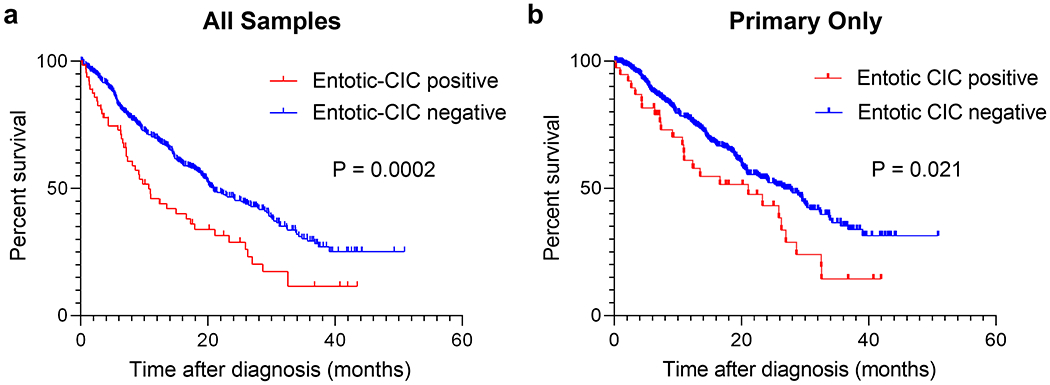
(a) Entotic-CIC positive PDAC (n = 65) showed poorer prognosis than negative PDACs (n = 434) (P = 0.0002, log-rank (Mantel-Cox) test) in all samples. (b) Entotic-CIC positive primary PDAC (n = 39) showed poorer prognosis than negative primary PDACs (n = 350) (P = 0.021, log-rank (Mantel-Cox) test).
Table 5.
Entotic-CIC in PDAC and Patient Outcome (univariate analysis)
| Factor | P-value | Hazard ratio (95% confidence interval) | ||
|---|---|---|---|---|
| Clinicopathologic | Entotic-CIC | (positive vs negative) | < 0.001 | 1.813 (1.323-2.484) |
| Age | (>65 vs <=65) | 0.853 | 1.024 (0.8-1.31) | |
| Gender | (male vs female) | 0.010 | 1.380 (1.082-1.761) | |
| Location | (primary vs metastasis) | < 0.001 | 0.327 (0-0.423) | |
| Histology | (PDA vs ASQ) | 0.415 | 0.778 (0.425-1.424) | |
| Genetic | KRAS | (mutated vs WT) | 0.969 | 0.991 (0.646-1.521) |
| TP53 | (mutated vs WT) | 0.173 | 1.216 (0.918-1.611) | |
| CDKN2A | (mutated vs WT) | 0.094 | 1.259 (0.962-1.649) | |
| SMAD4 | (mutated vs WT) | 0.976 | 0.996 (0.746-1.329) | |
| CDKN2AP16INK4A | (mutated vs WT) | 0.384 | 1.141 (0.848-1.534) | |
| ARID1A | (mutated vs WT) | 0.395 | 1.179 (0.807-1.725) | |
| CDKN2AP14ARF | (mutated vs WT) | 0.310 | 1.199 (0.844-1.703) | |
| RNF43 | (mutated vs WT) | 0.382 | 0.772 (0.433-1.378) | |
| KDM6A | (mutated vs WT) | 0.828 | 1.062 (0.619-1.82) | |
| APC | (mutated vs WT) | 0.166 | 1.464 (0.854-2.51) | |
| CNA alteration | MYC | (amplified vs not amplified) | 0.883 | 0.941 (0.419-2.115) |
| AKT2 | (amplified vs not amplified) | 0.399 | 1.297 (0.709-2.371) | |
| CCNE1 | (amplified vs not amplified) | 0.503 | 0.678 (0.217-2.116) | |
| KRAS | (amplified vs not amplified) | < 0.001 | 5.480 (2.419-12.416) | |
| FGFR1 | (amplified vs not amplified) | 0.117 | 2.490 (0.796-7.788) | |
| CDKN2A | (deep deletion vs no deletion) | 0.010 | 1.631 (1.122-2.372) | |
| CDKN2AP16INK4A | (deep deletion vs no deletion) | 0.019 | 1.575 (1.077-2.303) | |
| CDKN2AP14ARF | (deep deletion vs no deletion) | 0.016 | 1.604 (1.091-2.358) | |
| CDKN2B | (deep deletion vs no deletion) | 0.216 | 1.291 (0.862-1.934) | |
| SMAD4 | (deep deletion vs no deletion) | 0.242 | 0.639 (0.302-1.353) |
Table 6.
Multivariate Analysis of Entotic-CIC in PDAC and Patient
| Factor | P-value | Hazard ratio (95% CI) | |
|---|---|---|---|
| Entotic-CIC | (present vs absent) | 0.014 | 1.527 (1.090-2.138) |
| Gender | (male vs female) | 0.005 | 1.431 (1.115-1.836) |
| Location | (primary vs metastasis) | < 0.001 | 0.360 (0-0.474) |
| TP53 | (mutated vs WT) | 0.328 | 1.153 (0.866-1.535) |
| CDKN2A | (mutated vs WT) | 0.217 | 1.197 (0.900-1.592) |
| APC | (mutated vs WT) | 0.409 | 1.184 (0.792-1.771) |
| KRAS | (amplified vs not amplified) | 0.020 | 2.742 (1.173-6.408) |
| FGFR1 | (amplified vs not amplified) | 0.944 | 0.959 (0.295-3.119) |
| CDKN2A | (deep deletion vs no deletion) | 0.245 | 1.382 (0.801-2.386) |
Table 7.
Multivariate Analysis of Entotic-CIC in PDAC (Primary Only) and Patient
| Factor | P-value | Hazard ratio (95% CI) | |
|---|---|---|---|
| Enototic-CIC | (present vs absent) | 0.018 | 1.668 (1.092-2.548) |
| Gender | (male vs female) | 0.181 | 1.225 (0.910-1.651) |
| TP53 | (mutated vs WT) | 0.060 | 1.435 (0.986-2.090) |
| CDKN2A | (mutated vs WT) | 0.254 | 1.217 (0.868-1.708) |
| APC | (mutated vs WT) | 0.106 | 1.825 (0.881-3.781) |
| KRAS | (amplified vs not amplified) | 0.010 | 8.242 (1.658-40.974) |
| FGFR1 | (amplified vs not amplified) | 0.384 | 2.420 (0.331-17.674) |
| CDKN2A | (deep deletion vs no deletion) | 0.415 | 0.711 (0.314-1.613) |
Discussion
These findings not only validate prior reports in rectal, head and neck and breast cancer and but provide a deeper understanding of the clinicopathologic relevance of entosis in PDAC. We find that entosis in PDAC is a correlate of aggressive biology and an independent prognostic factor by multivariate analysis. Moreover, metastases appear to have a greater number of entotic-CICs than primary tumors.
Our finding that entotic-CICs are more prevalent in poorly differentiated PDAC parallels prior observations in breast cancer 9. We extend this observation by showing that pancreatic adenosquamous carcinoma (ASC) also have a high number of entotic-CICs. This is consistent with our prior reports that squamous features in PDAC arise in the context of clonal evolution and that ASC has more entotic-CICs than poorly differentiated carcinoma10. Furthermore, our finding of an association of TP53 mutations with entotic-CIC parallels prior reports in lung cancer.5 However, we now clarify the lack of an association with TP53 hot spot mutations. While the mechanisms by which TP53 alterations contribute to entosis is unknown, one putative molecular mechanism is that loss of TP53 or delta-133TP53 expression increases extracellular ATP release and the consequent activation of purinergic P2Y2 receptors, which induces cell engulfment 15. Alternatively, TP53 activation may cause upregulation of Rnd3, which inhibits ROCK1 or RhoA activities directly or indirectly, leading to polarized activation of RhoA-actomyosin which drives cell internalization to form CIC structures 16.
The reason for an increased prevalence of entotic-CICs in metastatic PDAC, specifically liver metastases, also remains to be determined. However, we expect that the mechanisms are multifactorial. For example, a recent report showed TP53 inactivation and KRAS amplification are more frequent in metastasis 17. Our data confirms this phenomenon because of eight KRAS amplified samples, six were liver metastases and only two were primary carcinomas (P < 0.0001, chi square test with Yates correction). While PDAC metastases does not appear to have a genetic basis 18, subclonal evolution at the genetic level with respect to gene dosages may select for the entotic phenotype. We suspect the tumor microenvironment may also play a role. For example, while primary tumors are rich in stromal cells and poorly vascularized, the liver is by contrast a highly vascular tissue with little stromal response until the metastases exceeds a critical mass of cells 19. Thus, it is conceivable that changes in the microenvironment and available nutrients may select for the entotic phenotype within specific genetic contexts.
Poor prognosis in entotic-CIC high tumors that we report here have been previously reported in lung adenocarcinoma 5, head and neck squamous cell carcinoma (HNSCC) 7,8, rectal and anal carcinoma 8. However, given that only in HNSCC has entotic-CIC been shown to be an independent prognostic factor in HNSCC 7 , this is the first demonstration of entotic -IC as an independent prognostic factor in adenocarcinoma. While we cannot totally exclude the possibility of simple dimensional overlap or emperipolesis 20 in tumor cells reported previously, we believe that our simple methods to evaluate entotic-CIC can be applied to practical diagnostic pathology. We also expect that our findings will also have implications for understanding entotic-CIC in other solid tumor types.
Supplementary Material
Acknowledgements
We are grateful for Laura D. Wood and Ralph H. Hruban for sample collection and helpful discussions. We gratefully acknowledge the members of the Molecular Diagnostics Service in the Department of Pathology for MSK IMPACT.
Footnotes
Disclosure/Conflict of Interest
Christine A. Iacobuzio-Donahue receives research support from Bristol Myers Squibb. David S. Klimstra. is a consultant and equity holder to Paige.AI, consultant to Merck Pharmaceuticals, and receives royalties from UpToDate and the American Registry of Pathology. Eileen M. O’Reilly has research funding to MSK from Celgene/BMS, BioNTech, ActaBiologica, AstraZenica, Silenseed, Arcus and Gossamer and is a consult to Merck, Cytomx, BioLineRx, Targovax, Celgene/BMS and Loxo, Polaris and Rafael. All other authors declare no competing interests.
References
- 1.Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018;25:486–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Overholtzer M, Mailleux AA, Mouneimne G, Normand G, Schnitt SJ, King RW, et al. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell 2007;131:966–979. [DOI] [PubMed] [Google Scholar]
- 3.Riegman M, Bradbury MS, Overholtzer M. Population Dynamics in Cell Death: Mechanisms of Propagation. Trends in Cancer. 2019;5:558–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamann JC, Surcel A, Chen R, Teragawa C, Albeck JG, Robinson DN, et al. Entosis Is Induced by Glucose Starvation. Cell Rep. 2017;20:201–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mackay HL, Moore D, Hall C, Birkbak NJ, Jamal-Hanjani M, Karim SA, et al. Genomic instability in mutant p53 cancer cells upon entotic engulfment. Nat Commun. 2018;9:3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun Q, Luo T, Ren Y, Florey O, Shirasawa S, Sasazuki T, et al. Competition between human cells by entosis. Cell Res. 2014;24(11):1299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schenker H, Buttner-Herold M, Fietkau R, Distel L V. Cell-in-cell structures are more potent predictors of outcome than senescence or apoptosis in head and neck squamous cell carcinomas. Radiat Oncol. 2017;12:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwegler M, Wirsing AM, Schenker HM, Ott L, Ries JM, Büttner-Herold M, et al. Prognostic Value of Homotypic Cell Internalization by Nonprofessional Phagocytic Cancer Cells. Biomed Res Int. 2015; 359392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krajcovic M, Johnson NB, Sun Q, Normand G, Hoover N, Yao E, et al. A non-genetic route to aneuploidy in human cancers. Nat Cell Biol. 2011;13:324–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashi A, Fan J, Chen R, Ho Y, Makohon-Moore AP, Lecomte N, et al. A unifying paradigm for transcriptional heterogeneity and squamous features in pancreatic ductal adenocarcinoma. Nat Cancer. 2020;1:59–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial sloan kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): A hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagnostics. 2015;17:251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.WHO_Classification_of_Tumours_Editorial_Board, ed. Digestive System Tumours WHO Classification of Tumours, 5th ed. : WORLD HEALTH ORGANIZATION, 2019. [Google Scholar]
- 14.Sun Q, Cibas ES, Huang H, Hodgson L, Overholtzer M. Induction of entosis by epithelial cadherin expression. Cell Res. 2014;24:1288–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Syed Qasim Raza. Caractérisation des bases moléculaires de l’entose (Article in French). 2012.
- 16.Liang J, Niu Z, Yu X, Zhang B, Wang M, Ruan B, et al. Counteracting Genome Instability by p53-dependent Mintosis. bioRxiv. 2020;2020.01.16.908954. [Google Scholar]
- 17.Connor AA, Denroche RE, Jang GH, Lemire M, Zhang A, Chan-Seng-Yue M, et al. Integration of Genomic and Transcriptional Features in Pancreatic Cancer Reveals Increased Cell Cycle Progression in Metastases. Cancer Cell. 2019;35:267–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makohon-Moore AP, Zhang M, Reiter JG, Bozic I, Allen B, Kundu D, et al. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat Genet. 2017;49:358–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan S, Norgard RJ, Stanger BZ. Cellular plasticity in cancer. Cancer Discov. 2019;9:837–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia P, Wang S, Guo Z, Yao X. Emperipolesis, entosis and beyond: Dance with fate. Cell Res. 2008;18(7):705–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


