Figure 4. Multiple cytotoxic CD4+ T cell states are enriched and clonally expanded in bladder tumors, and possess lytic capacity against tumors.
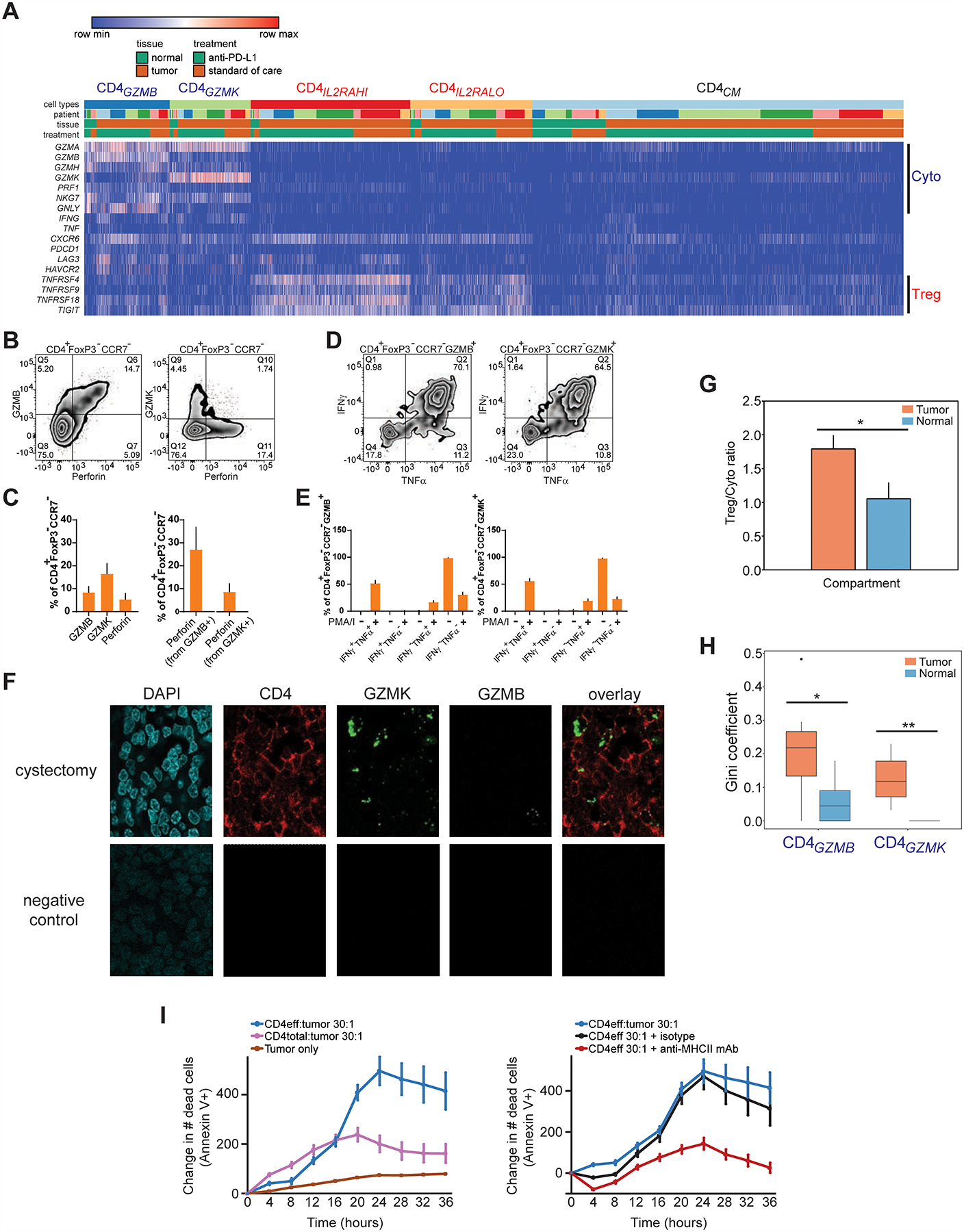
(A) Heatmap shows the expression of select cytotoxic or regulatory T cell marker genes (rows) for individual single cells (columns) within the cytotoxic CD4GZMB and CD4GZMK clusters, compared to regulatory (CD4IL2RAHI, CD4IL2RLO) and CD4CM clusters. Cells were grouped based on their annotations by tissue (tumor or non-malignant), treatment, and patient. Log2-transformed expression of each gene was row scaled. (B) Flow cytometry staining of GZMB, perforin, or GZMK in CCR7− CD4+ FOXP3− T cells is shown. (C) Percentage of cells expressing GZMB, GZMK, or perforin from CCR7− CD4+ FOXP3− T cells by flow cytometry (left), and the percentage of cells co-expressing perforin within GZMB+ or GZMK+ CCR7− CD4+ FOXP3− T cells (right), are shown (N = 7 tumors, mean + s.e.m.). (D) Representative flow cytometry staining of IFNγ and TNFα expression in GZMB+ or GZMK+ CCR7− CD4+ FOXP3− T cells stimulated with PMA and ionomycin. Bottom row, percentages of cells expressing IFNγ, TNFα, or both from GZMB+ or GZMK+ CCR7− CD4+ FOXP3− T cells with and without stimulation (N = 11 tumors, mean + s.e.m.). (F) Multiplex immunofluorescent staining of DAPI (blue), CD4 (immunohistochemistry, red), GZMK (RNAscope probe, green), GZMB (RNAscope probe, white) and overlay without DAPI are shown from a cystectomy tumor region from a patient with parallel scRNA-seq and TCR-seq data (anti-PD-L1 C, top row), and from a corresponding tumor field with negative control staining (bottom row). CD4+ cells that co-express GZMK (arrows) or GZMB (arrowhead) are indicated. Scale bar, 10 μm. (G) The ratio of abundances of all regulatory T cell populations (CD4ILRAHI, CD4IL2RALO) to all cytotoxic CD4+ populations (CD4GZMB, CD4GZMK) across all tumor and non-malignant samples is shown. (*, P < 0.05 by unpaired T test assuming unequal variance.) (H) Gini coefficients for each of the cytotoxic CD4+ populations within tumor and non-malignant compartments across all samples (*, P < 0.05, **, P < 0.01, exact permutation test. N = 7 tumor samples, 6 non-malignant samples.) (I) Left panel, quantitation of Annexin V+ apoptotic cells over time from a time-lapse cytotoxicity experiment with tumor cells cultured alone or with either bulk CD4+ TIL (CD4total) or CD4+ TIL depleted of regulatory T cells (CD4eff) at 30:1 effector:target ratio. Right panel, CD4eff TIL and tumor cells (30:1 effector:target ratio) were co-cultured with a pan-anti-MHCII antibody or isotype control. All traces were from the same culture and cytotoxicity assay from the same patient. All traces show relative change in cell death from timepoint 0. Cytotoxicity with CD4eff are representative of independent experiments from 4 different patients. Mean ± s.e.m. from multiple technical replicates for each experiment are shown.
