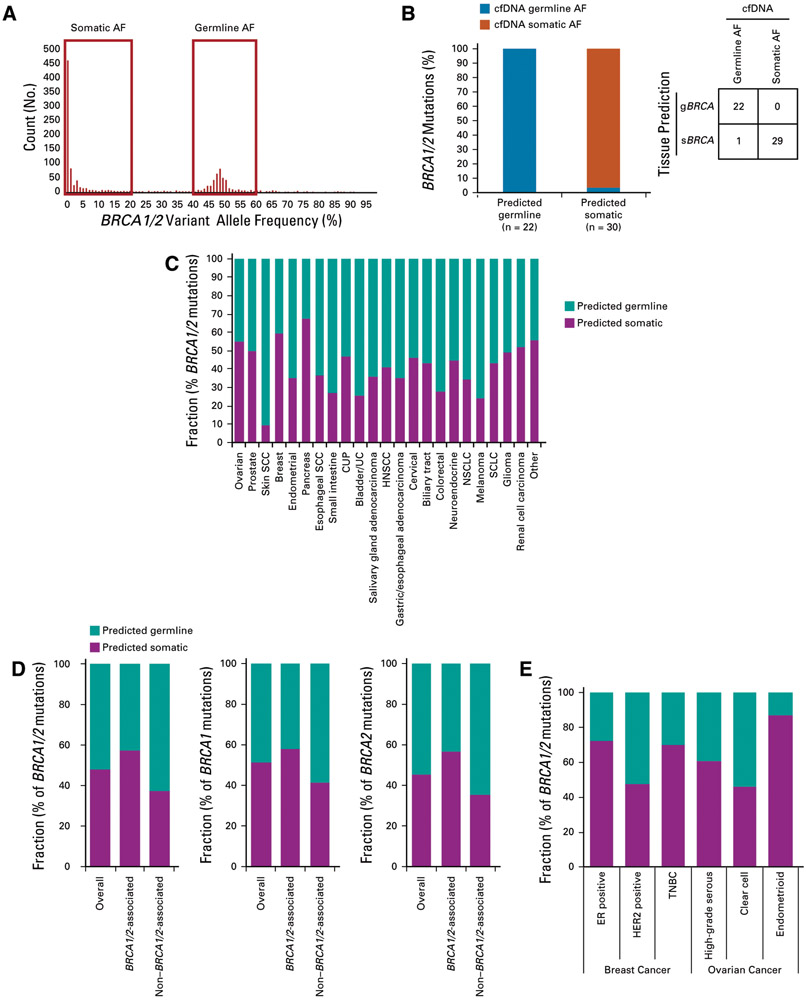
FIG A3.
(A and B) Comparison of tissue-based germline/somatic predictions to BRCA1/2 allele frequencies (AFs) in patient-matched cell-free DNA (cfDNA). (A) Distribution of AFs for 1,207 cfDNA samples with BRCA1/2 mutation. Mutations with somatic AFs were defined as those identified in cfDNA at 0%-20% AF. Mutations with germline AF were defined as those identified in cfDNA at 40%-60% AF, except for cases with high circulating tumor DNA fraction (20%) that were excluded from the analysis as ambiguous AF. (B) Fifty-two tissue-derived germline BRCA1/2 (gBRCA1/2) and somatic BRCA1/2 (sBRCA1/2) mutation predictions (from 48 tissue samples) were compared with patient-matched cfDNA AFs; 100.0% of germline predictions (22 of 22) were observed at germline-like AF and 96.7% of somatic predictions (29 of 30) were observed at somatic-like AF. (C-E) Predicted germline/somatic status calls were made for each BRCA1 or BRCA2 short variant mutation. For mutations yielding a successful call, frequency of predicted germline v somatic mutation was determined for (C) BRCA1/2 across cancer types, (D) BRCA1/2 (grouped and individually) overall (n = 5,845) for BRCA1/2-associated cancers (breast, ovarian, pancreatic, prostate; n = 3,061) and non–BRCA1/2-associated cancers (n = 2,784), and (E) BRCA1/2 for the subset of ovarian and breast cancer cases where molecular/histologic subtype information was available. CUP, cancer of unknown primary; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; HNSCC, head and neck squamous cell carcinoma; NSCLC, non–small-cell lung cancer; SCC, squamous cell carcinoma; SCLC, small-cell lung cancer; TNBC, triple-negative breast cancer; UC, urothelial cancer.