Extended Data Figure 1: Results from improved paralog-filtering.
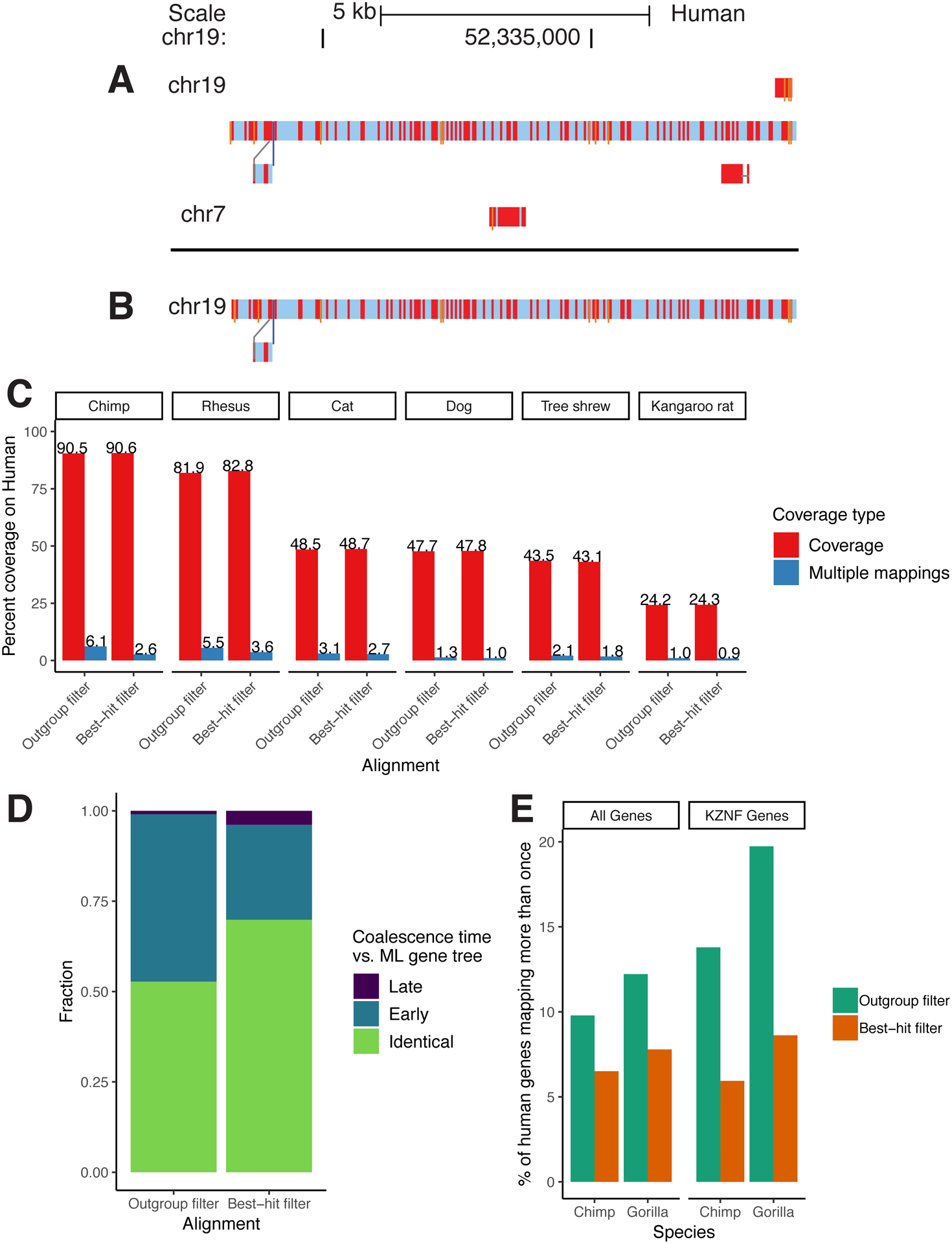
A/B: A sample snake track [45] within a recently duplicated region before (A) and after (B) the filtering change. Nucleotide substitutions are shown as red bars, and insertions are shown as thin orange bars. C: Coverage results from two alignments of identical assemblies using the outgroup and best-hit filtering methods. Multiple-mappings: sites which map to two or more sites on the target genome. D: Results from comparing phylogenetic trees implicit in the HAL alignment to ML re-estimated trees of the same regions. “Early” coalescences imply that too many duplication events have been created in the reconciled trees, while “Late” implies that too many loss events have been created. E: Percent of human genes that map more than once to the chimp/gorilla genomes in two CAT [9] annotations using alignments created with the outgroup and best-hit filtering methods. KZNF: KRAB zinc-finger genes.
