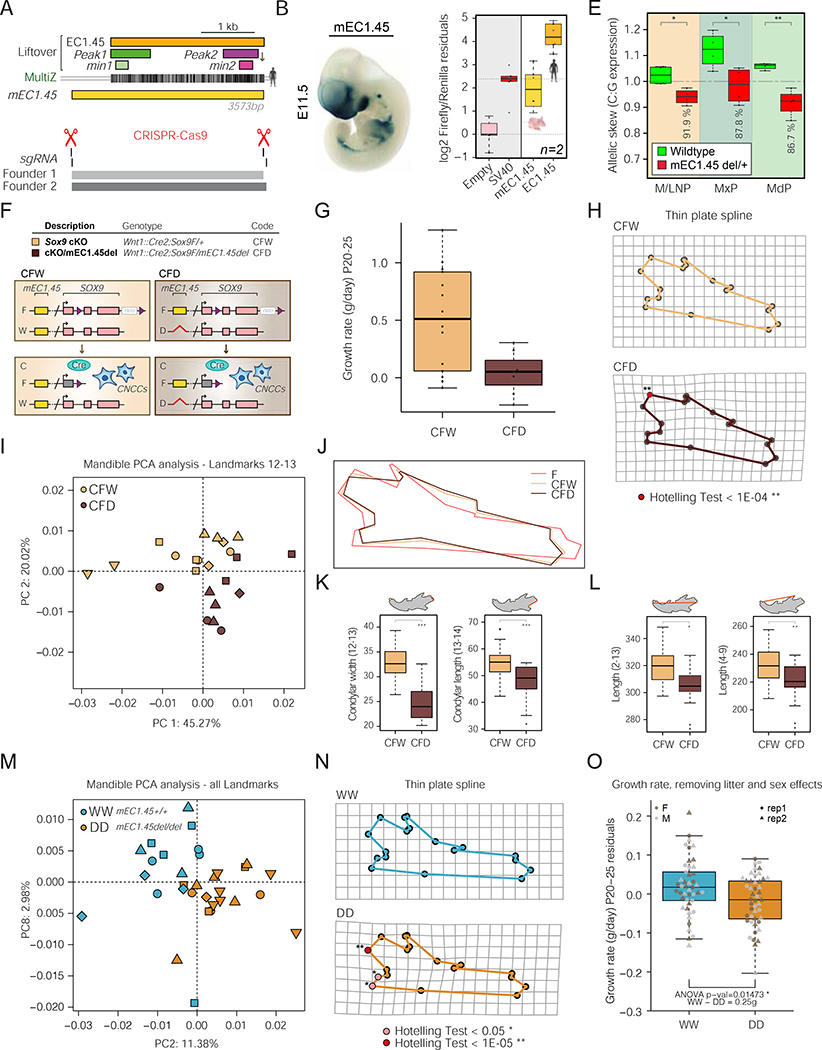
Figure 6: Reduction in Sox9 activity impacts mouse craniofacial development in a dosage-dependent manner.
(A) Schematic of mouse orthologous mEC1.45 with liftover of human EC1.45, Peak1, Peak2, min1 and min2 sequences, and human to mouse MultiZ alignment.
(B) Mouse lacZ reporter assay for mEC1.45 at E11.5.
(C) Luciferase assay for human EC1.45 and mouse mEC1.45.
(D) Location of sgRNAs and Founder 1 and 2 mEC1.45 deletions (aligned with A). (E) Digital droplet RT-PCR for Sox9 from wildtype and mEC1.45del/+ dissected E11.5 craniofacial tissues. Plotted as C:G allelic ratio, mEC1.45 deleted on C-allele. M/LNP, medial and lateral nasal processes; MxP, maxillary process; MdP, mandibular process. * pval < 0.05; ** pval < 0.01.
(F) Schematic of Sox9 heterozygous conditional knock-out Wnt1::Cre2;Sox9F/+ (CFW) and compound heterozygous Wnt1::Cre2;Sox9F/mEC1.45del (CFD) mice with Sox9 deleted in CNCCs on one allele, and mEC1.45 deleted on the other. Purple triangles, loxP sites; neo, neomycin resistance.
(G) Boxplot of postnatal growth rate (P20-P25, g/day) for CFW and CFD animals, ANOVA p-val = 0.01676.
(H) Landmarks for CFW (upper) and CFD (lower) mandibles projected onto a thin plate spline. Landmarks that differ significantly by Hotelling test are highlighted in red (p < 1E-04).
(I) PCA plot of mandible landmarks 12–13 following Procrustes analysis at E18.5 for 5 litters (23 embryos) of CFW (yellow) and CFD (brown) embyros.
(J) Procrustes transformed average mandible wireframes for wildtype (dark pink, FW), CFW (yellow) and CFD (brown) embryos.
(K) Measurements of width and length of the condylar process for CFW (yellow) and CFD (brown) mandibles. For condylar width, ANOVA p-val = 1.52E-07 and for condylar length, ANOVA p-val = 1.11E-04.
(L) As for K, two measurements of mandible length (2–13), ANOVA p-val = 0.00143 and (4–9), ANOVA p-val = 0.00687.
(M) PCA plot of all mandible landmarks following Procrustes analysis for wildtype (mEC1.45+/+, blue, WW) and homozygous mutant (mEC1.45del/del, orange, DD) embryos at E18.5 for 5 litters (32 embryos).
(N) Landmarks for WW and DD mandibles projected onto a thin plate spline. Landmarks that differ significantly by Hotelling test are highlighted in pink (p < 0.05) and red (p < 1E-05).
(O) Boxplot of postnatal growth rate (P20-P25, g/day) for WW and DD embryos. Two replicate groups plotted as residuals of linear regression, ANOVA p-val = 0.01473.
For PCA plots, different shape markers represent independent litters.