Fig 1.
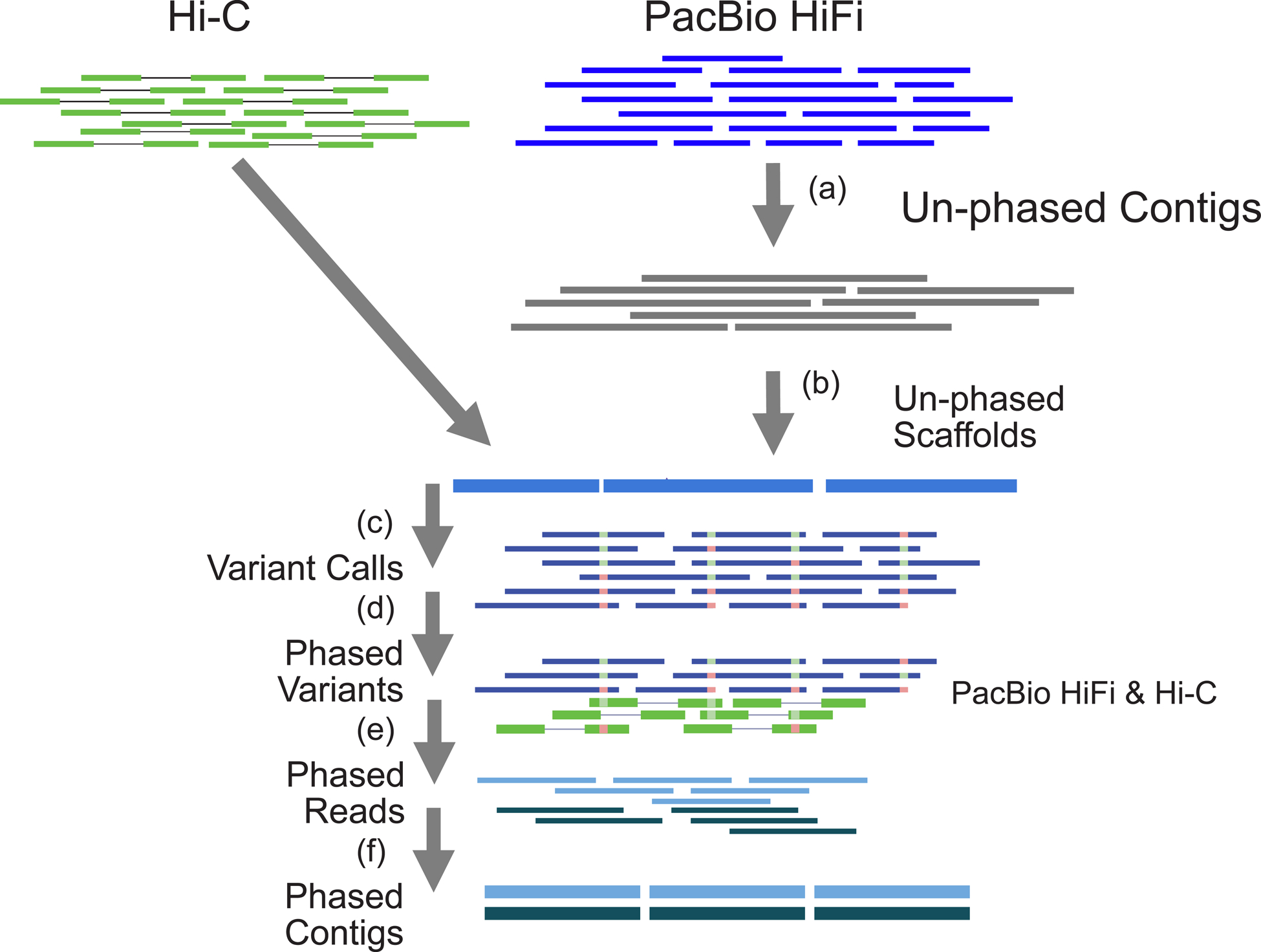
Outline of the phased assembly algorithm: DipAsm. (a) Assemble HiFi reads into unphased contigs using Peregrine. (b) Group and order contigs into scaffolds with Hi-C data using HiRise/3D-DNA. (c) Map HiFi reads to scaffolds and call heterozygous SNPs using DeepVariant. (d) Phase heterozygous SNP calls with both HiFi and Hi-C data using WhatsHap+HapCut2. (e) Partition reads based on their phase using WhatsHap. (f) Assemble partitioned reads into phased contigs using Peregrine.
