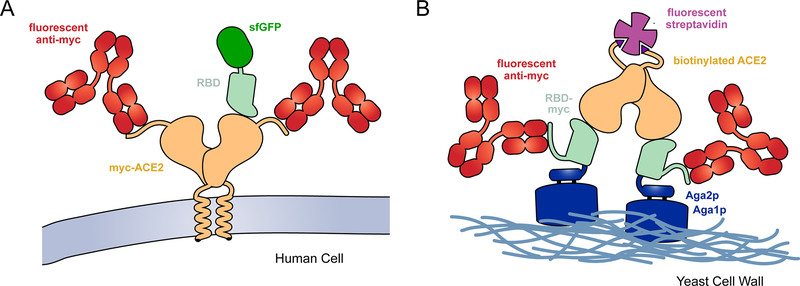
Figure 1. Selection strategies for deep mutational scans of the RBD•ACE2 complex.
(A) A library of ACE2 variants was expressed in human cells. Full-length ACE2 (tan) was tagged with a c-myc epitope at its extracellular N-terminus for detection of surface expression with a fluorescent antibody (red). SARS-CoV-2 RBD (pale green) genetically fused with superfolder green fluorescent protein (sfGFP; dark green) was incubated with the cell culture. FACS was used to collect fluorescent cells expressing ACE2 with bound RBD-sfGFP. (B) The isolated RBD of SARS-CoV-2 (pale green) was fused at its N-terminus to Aga2p (blue) and at its C-terminus to a c-myc epitope tag. A saturation mutagenesis library of the RBD was expressed on the yeast surface. Following induction of RBD expression, the yeast were incubated with dimeric, biotinylated sACE2 (tan). Bound ACE2 was detected with fluorescent streptavidin (purple) and surface expressed RBD was detected with a fluorescent antibody (red).