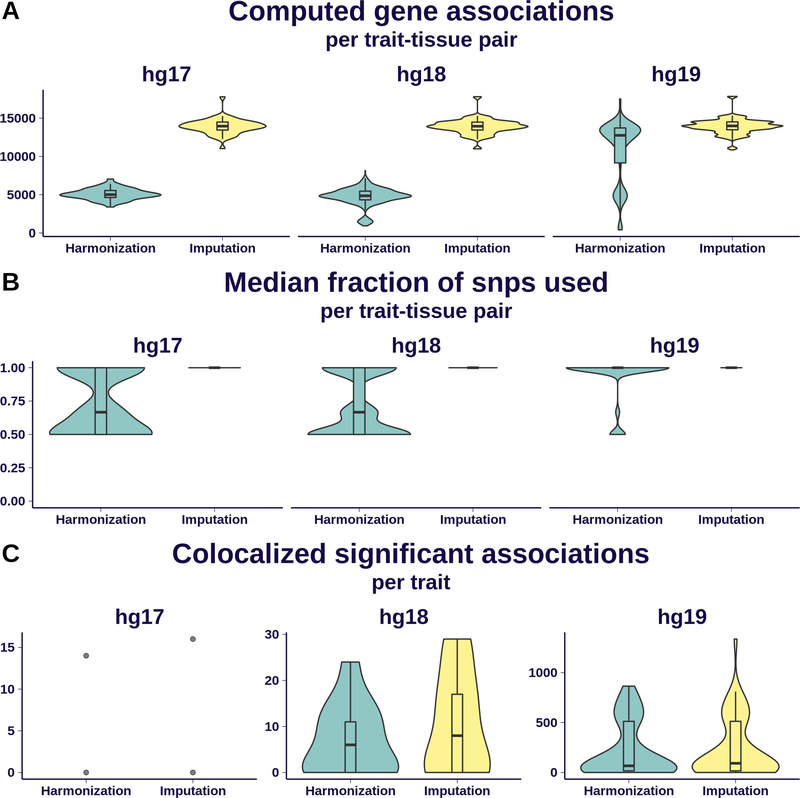
Fig 5. Effect of imputation on association quality.
We display here a comparison of S-PrediXcan results from MASHR-M models on 69 GWAS traits using two different pre-processing schemes: simple harmonization of GWAS variants to GTEx’s, and additional imputation of missing summary statistics. Results are grouped by the different human genome release versions underlying each GWAS: 2 traits were defined on hg17, 13 on hg18, and 54 on hg19.
Panel A shows the distribution of number of associations per trait-tissue pair that can be computed; imputation dramatically increased the number of associations for hg17- and hg18-based traits. Some hg19-based traits exhibited a good number of computable associations after just a simple harmonization.
Panel B shows, per trait-tissue pair, the distribution of median fraction of model SNPs present in the GWAS. It is nearly 1 for most trait-tissue pairs in the imputation scheme, ranging between 0.5 and 1 with the harmonization scheme.
Panel C shows the number of colocalized, significantly associated genes that can be found after applying imputation and harmonization schemes. The gain of imputation for hg19 is less dramatic that in the other comparisons in this figure, given the conservative nature of the colocalization filter.