Figure 3. Performance of deep learning models trained to predict BRS.
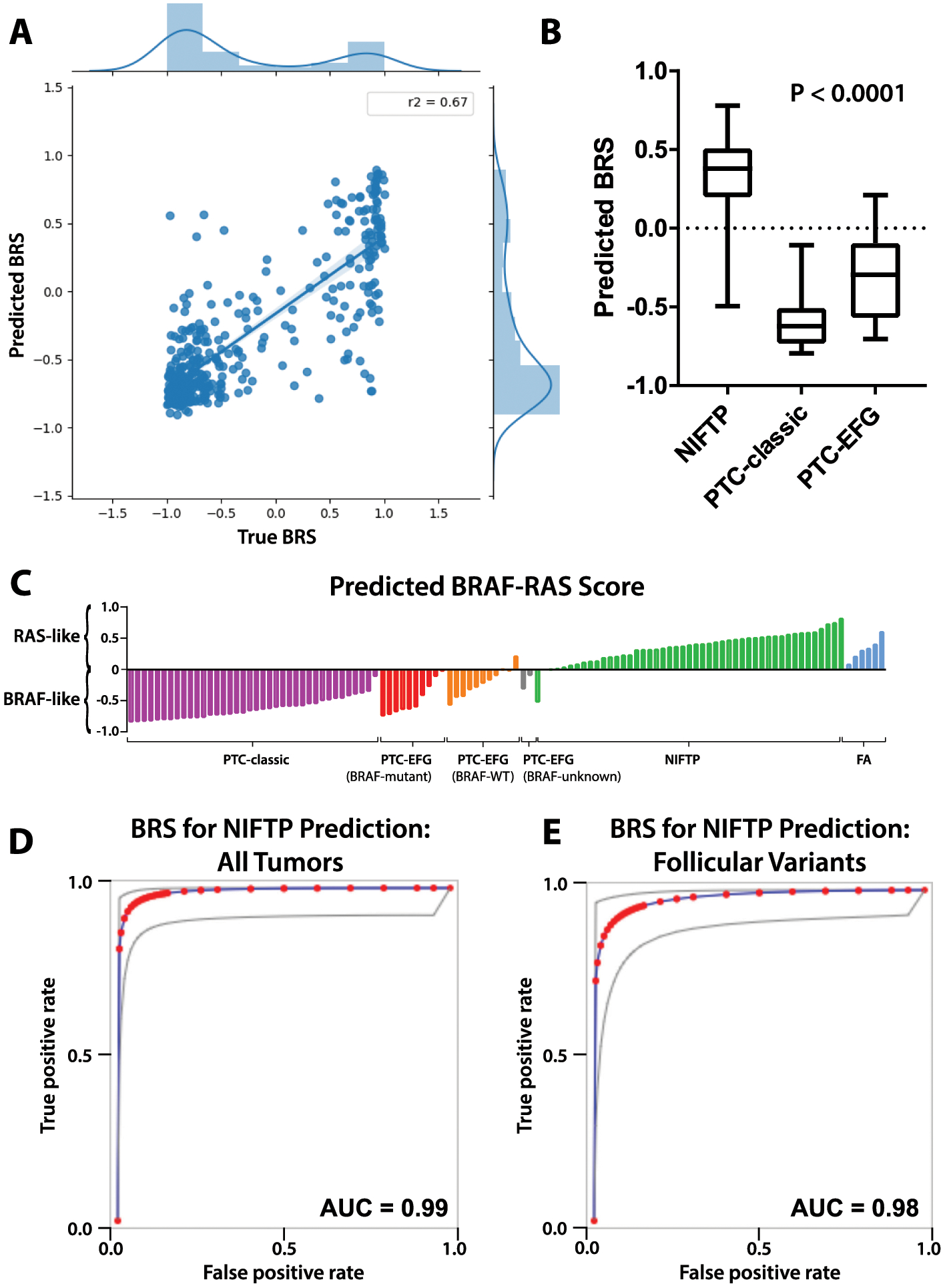
A. Xception-based deep learning models were trained on the TCGA thyroid cohort to predict BRS as a linear outcome using three-fold cross validation. For each slide in the the left-out validation sets, slide-level score predictions were generated by averaging predictions across constituent tiles. Predicted BRS was plotted against true BRS for each k-fold, with R2 calculated 0.67, 0.73, and 0.61 for each of the k-folds. The three plots were then aggregated across k-folds as shown in A, with an aggregated R2 of 0.67. B. A final model was generated by training across the entire TCGA thyroid cohort (DL-TCGA-BRS) and was used to predict BRS for slides in the institutional dataset. Distribution of BRS predictions can be seen across NIFTP, PTC-classic, and PTC-EFG slides. NIFTP tumors had significantly higher predicted BRS, indicating a more RAS-like phenotype, than either of the PTC subclasses, as calculated using ANOVA (p < 0.0001). C. BRS predictions across slides in the institutional dataset, separated by tumor subtype. NIFTPs are near universally predicted to be RAS-like, with PTC-classics predominantly BRAFV600E-like. PTC-EFG tumors are largely BRAFV600E-like but demonstrate somewhat greater heterogeneity. D. Predicted BRS is an excellent discriminator of NIFTP status; using predicted BRS as a surrogate for NIFTP prediction, the test has an AUC of 0.99 when applied across all PTC-classic, PTC-EFG, and NIFTP tumors in the institutional dataset. E. Used as a discriminator solely between PTC-EFG and NIFTP tumors, predicted BRS has an AUC of 0.98.
