Figure 3. N-terminal acetylation in NAA15+/− and NAA15−/− iPSCs.
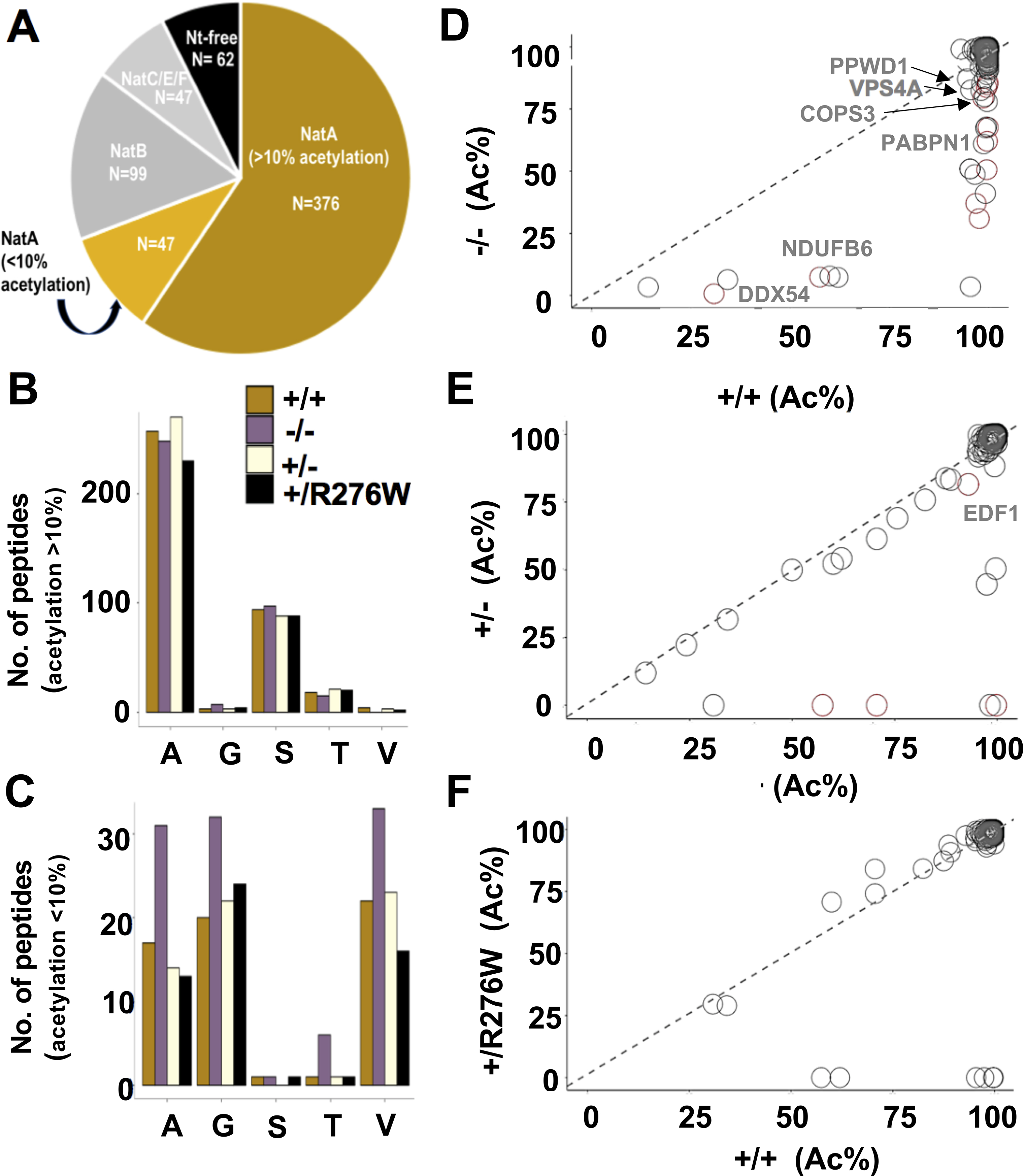
A) N-terminal peptides acetylated in NAA15+/+ iPSCs are categorized by NAT-type substrate specificity. NatA targets are displayed according to percent Nt-acetylation; Nt-acetylation >10% highlighted in gold and Nt-acetylation <10% highlighted in light gold. B-C) The number of Nt-acetylated NatA-type N-terminal peptides represented by residues A, G, S, T, or V in NAA15 mutant and wildtype iPSCs. Data is displayed as the absolute number of peptides measured from the sum of two biological replicates for each genotype. Alanine and serine residues are preferentially acetylated in peptides with >10% Nt-acetylation; glycine and valine residues are representative of N-termini with <10% Nt-acetylation. Primary peptide summary provided in Online Table V. There were no statistically significant differences between genotypes (hypergeometric test; data not shown). D-F) Representative scatter plots display the correlation of the degrees of Nt-acetylation of all N termini identified as NatA targets in NAA15−/−, NAA15+/−, NAA15+/R276W and NAA15+/+ iPSCs. Each plot compares NAA15 mutant iPSCs to wildtype iPSCs. Differentially acetylated N-termini are found below the line of identity (y intercept = 0, slope =1). Peptides lose acetylation moieties in NAA15-mutant iPSCs. NAA15−/− iPSCs have the largest number of peptides (n=32) with reduced Nt-acetylation. There are N-termini with reduced N-terminal acetylation in NAA15+/− iPSCs (n=9) and NAA15+/R276W iPSCs (n=8). Decreased protein expression changes are highlighted in red.
