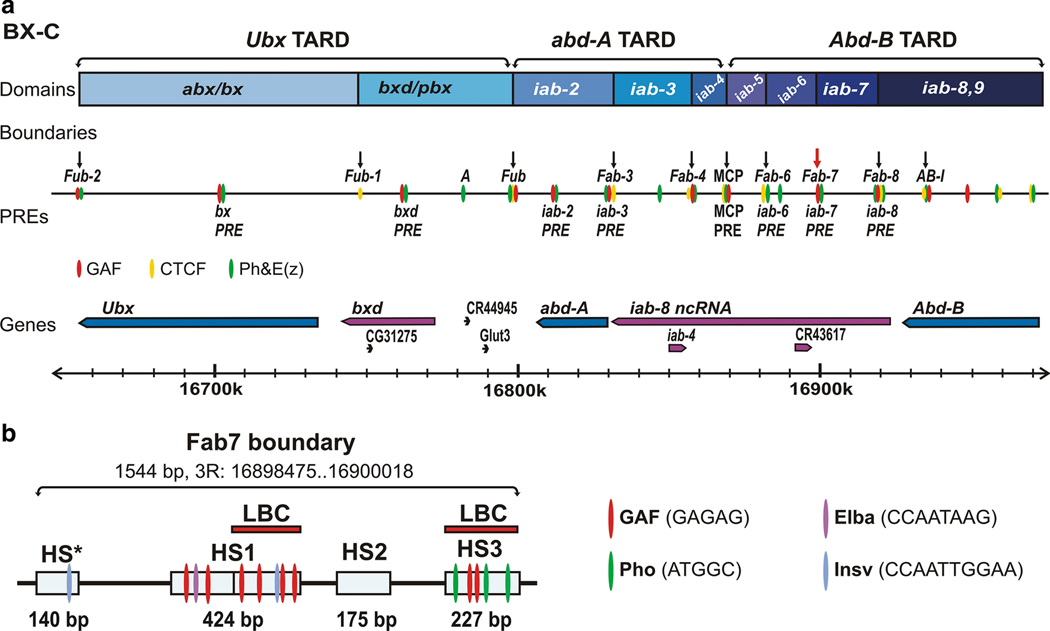
Fig. 4. GAF is required for the functioning of chromatin boundaries and PREs in BX-C.
(a) The diagram shows the BX-C TARDs, containing Ubx, abd-A, and Abd-B genes and their respective cis-regulatory domains. The positions of the boundaries and PREs are indicated above and below the sequence coordinate line correspondingly. The red arrow marks Fab7 boundary. The red, yellow and green ovals are regions bound by GAF, CTCF or E(z) and Ph, respectively. (b) The Fab-7 boundary has four nuclease hypersensitive regions: one minor - HS* and three prominent - HS1, HS2, and HS3. The locations of the recognition motifs for GAF, Pho, Elba and Insv proteins known to be associated with Fab-7 are indicated. The LBC complex binds to distal half of HS1 (dHS1) and HS3.