Figure 1: Generation of Marker-free (MF) lettuce plants expressing dextranase, mutanase and lipase and evaluation of transgene integration, marker removal and homoplasmy.
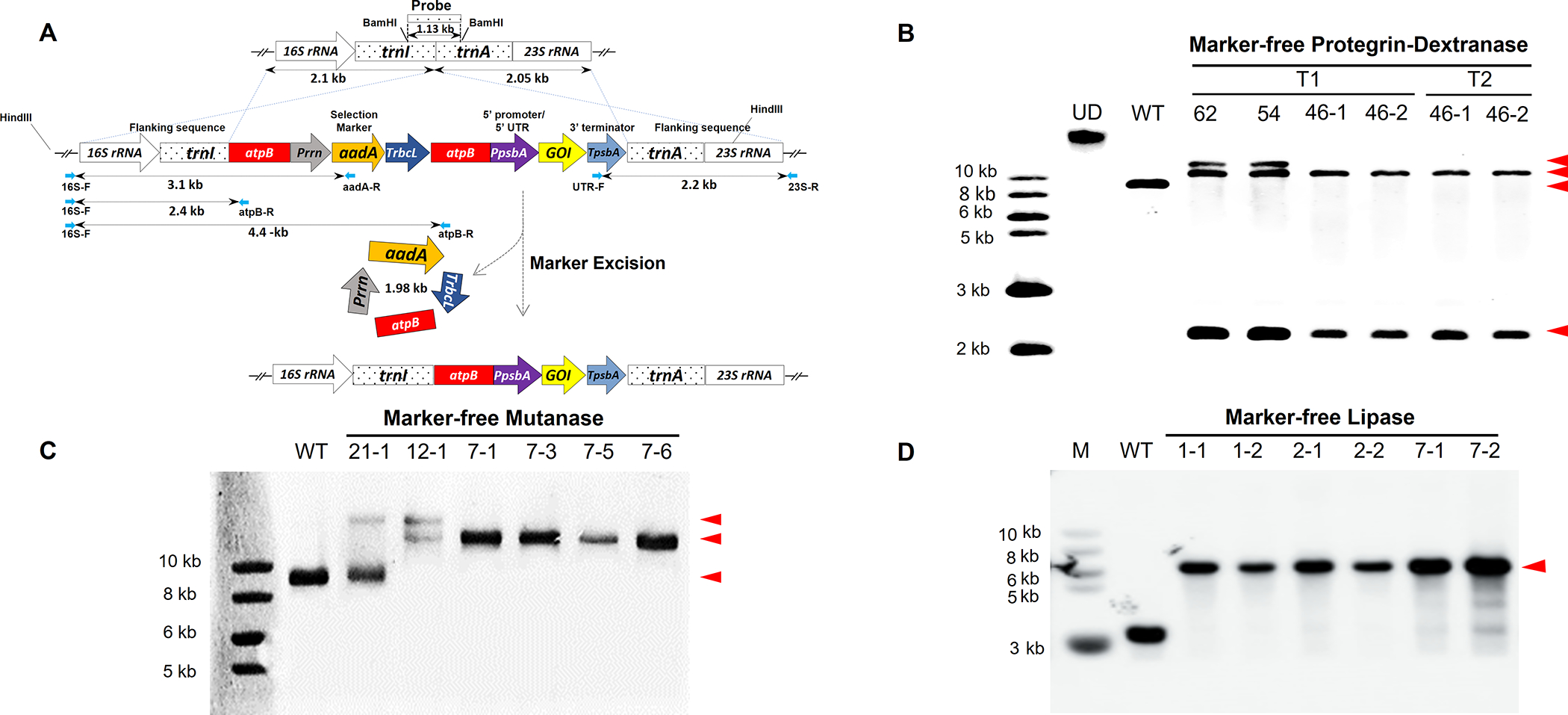
Schematic representation of the integration of two expression cassettes (gene of interest - GOI and selectable marker) into lettuce chloroplast genome via homologous recombination of flanking sequences: 16S rRNA-trnI and tranA-23S rRNA and subsequent removal of the antibiotic resistance gene via homologous recombination between two identical atpB regions. GOI represents PG1-Smdex or mut or lipY. Probe indicates the DNA fragment region which was digested by Bam HI and used to detect hybridizing fragments in Southern blots. (A). Southern blots confirm PG1-Smdex gene integration, marker removal and homoplasmy in transplastomic plants with 10.5 kb with 2.2 kb fragments, while 12.5 kb with 10.5 kb and 2.2 kb demonstrated heteroplasmy (with or without the aadA gene) after gDNA was digested with HindIII. Untransformed plant (WT) and undigested DNA (UD) were used as controls (B). In MF mutanase T0 generation, expected bands of 14.1 or 16.1 kb as a result of HindIII digested gDNA confirms mut gene integration in T0 generation plants, and the 14.1 kb band alone represents the homoplasmy (C). Expected band size of 5.6 kb obtained from SmaI digested gDNA confirms lipY gene integration, antibiotic marker gene removal and homoplasmy in lipase expressing T1 generation plants (D). Gene of interest band size is represented with arrowheads.
