Abstract
Recent evidence has highlighted the role of N6-methyladenosine (m6A) in the regulation of mRNA expression, stability and translation, supporting a potential role for post-transcriptional regulation mediated by m6A in cancer. Here we explore prostate cancer as an exemplar and demonstrate that low levels of N6-adenosine-methyltransferase (METTL3) is associated with advanced metastatic disease. To investigate this relationship, we generated the first prostate m6A maps, and further examined how METTL3 regulates expression at the level of transcription, translation, and protein. Significantly, transcripts encoding extracellular matrix proteins are consistently upregulated with METTL3 knockdown. We also examined the relationship between METTL3 and androgen signaling and discovered the upregulation of a hepatocyte nuclear factor-driven gene signature that is associated with therapy resistance in prostate cancer. Significantly, METTL3 knockdown rendered the cells resistant to androgen receptor antagonists via an androgen receptor independent mechanism driven by the upregulation of nuclear receptor NR5A2/LRH-1.
Implications:
These findings implicate changes in m6A as a mechanism for therapy resistance in metastatic prostate cancer.
Keywords: m6A, epitranscriptomics, prostate cancer, METTL3, castration resistance
INTRODUCTION
At the molecular level, prostate cancer (PCa) is generally characterized by large-scale structural genomic rearrangements1,2 and relatively few recurrent genomic alterations3,4, which thus far are insufficient to explain the heterogeneity of the disease, and suggest the importance of other mechanisms for gene regulation. In particular, multi-omics studies in PCa have demonstrated that for many oncogenes copy number alterations, DNA methylation, and even mRNA abundance are insufficient to explain the variation seen in protein expression5,6, emphasizing a sizeable role for post-transcriptional gene regulation in PCa.
Analogous to epigenetic modifications of DNA, mRNA is also subject to multiple biochemical modifications, the most prevalent being N6-methyladenosine (m6A)7. m6A is a dynamic mRNA modification and is regulated by the methyltransferase “writer” complex (METTL3, METTL14, WTAP, VIRMA, ZC3H13 and RBM15)8-12 the demethylase “eraser” (ALKBH5)13 and “reader” proteins (YTHDF1/2/3)14,15. m6A is found on ~25% of all transcripts, and is unevenly distributed, with the majority of residues localized near the stop codon or in 3’ untranslated regions (UTRs). Mechanistically, m6A has been shown to both destabilize mRNA transcripts9,15,16 and increase translation17, in addition to influencing splicing14,18 and miRNA processing19.
Alterations in m6A have been associated with progression of multiple cancer types. In glioblastoma and breast cancer, increased expression of the demethylase ALKBH5 and concurrent reduced levels of m6A were implicated in the promotion of tumorigenesis and self-renewal of stem-like cells by stabilizing key mRNAs (e.g. the pluripotency factor NANOG)20-22. In contrast, in lung adenocarcinoma and myeloid leukemia, elevated expression of METTL3 promoted the translation efficiency of several important oncogenes, and enhanced cell growth and invasion23,24. Lastly, METTL3 has been shown to be both highly expressed in prostate cancer, and essential for proliferation in multiple prostate cancer cell lines25-27, strongly supporting the significance of m6A methylation in PCa.
Of particular concern clinically is metastatic, castration-resistant PCa (CRPC) in which the disease develops resistance to both first-line treatment with androgen deprivation therapy, and to second generation androgen receptor (AR) signaling inhibitors (ARSi, ie. abiraterone or enzalutamide)28. There are several proposed mechanisms of resistance that lead to CRPC including alterations in AR which permit activity in low-androgen settings, alterations in pathways downstream of AR, and increased signaling through other signaling pathways allowing for AR independence29. Despite some recent progress in developing new treatments for CRPC, average survival at this advanced disease stage is approximately 3 years30. Determining the mechanisms behind ARSi resistance are essential to improve existing therapies and finding new potential drug targets.
To address this gap in knowledge, we investigated the role of m6A and METTL3 in the progression of PCa, with a particular focus on its role in regulating AR signaling and the response to ARSi. We show that low levels of METTL3 in patients is associated with clinical markers of CRPC, in particular dysregulation of AR signaling. Using miCLIP we mapped which transcripts are marked by m6A, and then further examined how METTL3 knockdown alters expression at the level of transcript, translation and protein. In comparing these cell line results to patient data we find that low METTL3 consistently increases expression of extracellular matrix genes. Furthermore, we examined the relationship between METTL3 and AR signaling and show that while knockdown of METTL3 has no effect on AR response genes, it does render the cells resistant to ARSi in an AR-independent manner that is driven by the upregulation of NR5A2. Overall, these findings support a new model for m6A function in affecting therapeutic sensitivity to ARSi, and suggests that patients with low levels of METTL3 expression may not demonstrate an optimal response from ARSi.
METHODS
Cell Lines
LNCaP cells (male, ATCC, RRID: CVCL_1379) were maintained in RPMI medium (Gibco, A1049101), supplemented with 10% FBS (Gibco, 10270106), and 1% penicillin-streptomycin (Gibco, 11548876) on poly-L-lysine coated plates. RWPE cells (male, ATCC, RRID: CVCL_3791) were maintained in Keratinocyte Serum Free Medium (Gibco, 17005075) supplemented with bovine pituitary extract and human recombinant EGF (included), and 1% penicillin-streptomycin (Gibco, 11548876). HEK293T cells (female, ATCC, RRID: CVCL_0063) and DU145 cells (male, ATCC, RRID: CVCL_0105) were maintained in DMEM (Gibco, 31966021), supplemented with 10% FBS, and 1% penicillin-streptomycin. All cell lines were grown at 37 °C with 5% CO2. All cell lines were authenticated by STR analysis and regularly tested for mycoplasma.
m6A individual-nucleotide-resolution cross-linking and immunoprecipitation (miCLIP)
Total RNA from LNCaP and RWPE cells was isolated with TRIzol according to the manufacturer instructions. PolyA+ mRNA was isolated using the PolyATtract mRNA Isolation System (Promega, Z5210). 20 μg of mRNA was used as input for miCLIP following the previously reported protocol 31 and an m6A antibody from Abcam (ab151230; RRID: AB_2753144). In parallel total mRNA was subjected to library preparation using the Illumina library preparation protocol. Final libraries were sequencing on an Illumina HiSeq2500 generating 50 bp paired-end reads at the Weill Cornell Medicine Epigenetic Core facility.
Generation of inducible knock-down lines
Tet-pLKO-puro was a gift from Dmitri Wiederschain (Addgene plasmid # 21915; http://n2t.net/addgene:21915 ; RRID: Addgene_21915). Tet-pLKO-puro was digested with AgeI and EcoRI and ligated with annealed oligos (Supplementary Table 1). Lentivirus was produced in HEK293T cells by transfection with the pLKO constructs, and subsequent virus containing media was used to transduce LNCaP cells. Three days post transduction the cells were subjected to puromycin selection (1 μg/mL).
RNA- and Ribo-Seq
For paired RNA and Ribosome profiling experiments cells (~15x106) were incubated with 100 μg/ml cycloheximide for 5 minutes at 37 °C. Cells were then trypsinized, pelleted, and the pellets washed twice with ice-cold PBS containing 100 μg/ml cycloheximide. An aliquot was set aside for confirmation of knock-down by western blot, and the remaining cells were resuspended in 425 μl of hypotonic buffer (5 mM Tris-HCl (pH 7.5), 2.5 mM MgCl2, 1.5 mM KCl and 1x Halt protease inhibitor cocktail (Thermo Scientific, 78410)), followed by the addition of 50 μg cycloheximide, 1 μl of 1M DTT, and 100 units of RNAse Inhibitor (Thermo Fisher Scientific, N8080119). The pellet was vortexed, 25 μl 10% Triton X-100 and 25 μl 10% sodium deoxycholate were added then followed by another vortex. The lysates were then cleared by spinning at 16,000 g for 8 minutes at 4 °C. The lysate was diluted 1:10 and OD at 260 nm was determined using a Nanodrop spectrophotometer.
Ribosome footprinting was based on the manufacturer’s instructions for the TruSeq Ribo Profile (Mammalian) Library Prep Kit (Illumina, discontinued). Briefly, lysates were normalized to OD of 30 and a volume of 200 μl. Then 3 μl of RNAse I (Ambion, AM2294) was added and the samples were incubated for 45 minutes at room temperature with shaking. 10 μl of SUPERase•In was added to stop the reaction. Ribosome bound RNA fragments were purified on MicroSpin S-400 HR Columns (Cytiva, 27-5140-01) followed by cleanup with the RNA Clean & Concentrator-25 kit. rRNA was removed using the NEBNext rRNA Depletion Kit (NEB, E6310), and 28-30 bp footprints were purified on a 15% polyacrylamide TBE/Urea gel. Ribo-seq libraries were then prepared with the purified footprints and the SMARTer Small RNA-Seq Kit (Takara, 635029) following the manufacturer’s instructions.
RNA was extracted from either 100 μl of ribosome footprinting lysate with the RNA Clean & Concentrator-25 kit (Zymo, R1017), or directly from cells using the RNeasy Mini Kit (Qiagen, 74106), and genomic DNA was removed using the DNA-free kit (Ambion, AM1906). RNA quality was assessed with a BioAnalyzer and quantity on a Qubit fluorometer. RNA-seq libraries were prepared using 1 μg of RNA and the NEBNext Ultra™ II RNA Library Prep Kit (NEB, E7775) with rRNA depletion following the manufacturer’s instructions.
All libraries were sequenced on an Illumina NextSeq generating 75 bp single-end reads at University of Basel Visceral Surgery and Precision Medicine Research Laboratory.
Bioinformatics Analysis of Sequencing Data
miCLIP
Crosslinking-induced mutation sites were identified in miCLIP datasets as described previously 31. Individual m6A sites were subjected to metagene analysis using MetaPlotR 32. These identified sites were further filtered only reporting those sites that had greater than one crosslinking-induced mutation (m ≥ 2) and those that mapped to an A residue to generate Supplementary Data 1.
RNA-Sequencing
Sequence reads were aligned to the human reference genome GRCh37 by STAR (RRID:SCR_004463) using the two-pass approach 33. Transcript quantification was performed using RSEM (RRID:SCR_013027) 34. Genes without >10 counts in at least 2 samples were discarded. Counts were normalized using the median of ratios method from the DESeq2 (RRID:SCR_000154) package in R version 3.6.1 (R Core Team (2019). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.). Differential expression analysis was performed using the wald test in DESeq2 35.
Ribo-Sequencing
Sequence reads were aligned as for RNA-seq, using STAR, but with the options – "winAnchorMultimapNmax 200 --seedSearchStartLmax 15 --outFilterMultimapNmax 20”, to improve mapping of short Ribo-seq reads. Differential analysis of ribosome loading was performed using Riborex which uses generalised linear models and the DESeq2 normalisation framework to identify genes showing altered translation between sample groups 36.
Drug Treatments
For all drug treatments cells were pre-treated with 200 ng/ml doxycycline for 96 hours to induce shRNAs. For DHT stimulation experiments cells were starved of hormone for 48 hours in phenol red-free RPMI media (Gibco, 11-835-030) with 10% charcoal stripped FBS (Gibco, A3382101), then treated with 10 nM dihydrotestosterone or 10 μM enzalutamide for 24 hours.
For Incucyte experiments cells were treated with doxycycline for 96 hours and then plated in a 96-well plate, 5000 cells per well, n = 8 per condition. Cell confluency was determined using the Incucyte S3 instrument and the IncuCyte S3 2018B software. Values were calculated as fold-change in confluency as compared to vehicle treated controls.
Double Knock-Down Experiments
Inducible shRNA lines were pre-treated with doxycycline for 72 hours and then plated in a 96-well plate, 10000 cells per well while continuing doxycycline treatment. The next morning cells were transfected with SMARTpool (Dharmacon) pools of 4 siRNAs targeting AR (L-003400-00-0005), NR5A2 (L-003430-00-0005), or a non-targeting pool (D-001810-10-05). That evening (8 hours after transfection) cells were treated with media containing 10 μM enzalutamide or DMSO, n = 4 per condition and placed in the Incucyte.
Proteomics
Inducible shRNA lines were treated with 200 ng/mL doxycycline for 96 hours. Cells were scraped directly in lysis buffer (8M urea, 100mM Tris pH8, plus protease inhibitors) and lysed with sonication for 1 minute on ice with 10 seconds intervals. The supernatant was reduced, alkylated and precipitated overnight. The pellet was re-suspended in 8M urea/50mM Tris pH8 and protein concentration was determinate with Qubit Protein Assay (Invitrogen).
10 μg of protein were digested with LysC for 2 hours at 37 °C followed by Trypsin at room temperature overnight. 800 ng of digests were loaded in random order onto a pre-column (C18 PepMap 100, 5 μm, 100 A, 300 μm i.d. x 5 mm length, Thermo Fisher, 160454) at a flow rate of 50 μL/min with solvent C (0.05% TFA in water/acetonitrile 98:2).
After loading, peptides were eluted in back flush mode onto a home packed analytical Nano-column (Reprosil Pur C18-AQ, 1.9 μm, 120 A, 0.075 mm i.d. x 500 mm length) using an acetonitrile gradient of 5% to 40% solvent B (0.1% Formic Acid in water/acetonitrile 4,9:95) in 180 min at a flow rate of 250 nL/min. The column effluent was directly coupled to a Fusion LUMOS mass spectrometer (Thermo Fischer, Bremen; Germany) via a nano-spray ESI source.
Data acquisition was made in data dependent mode with precursor ion scans recorded in the orbitrap with resolution of 120’000 (at m/z=250) parallel to top speed fragment spectra of the most intense precursor ions in the Linear trap for a cycle time of 3 seconds maximum.
The mass spectrometry data were processed with MaxQuant 37 (version 1.6.1.0) against the Swissprot Homo Sapiens database 38 (release February 2019). The initial precursor mass tolerance was set to 10 ppm, and that of the fragment peaks to 0.4 Da. Enzyme specificity was set to strict trypsin, and a maximum of three missed cleavages were allowed. Carbamidomethylation on cysteine was set as a fixed modification, methionine oxidation and protein N-terminal acetylation as variable modifications. The same fraction number was given to all replicates of a group, but each group was given a fraction number differing by 3, so that match between runs were prevented between the different groups.
Protein intensities are reported as MaxQuant’s Label Free Quantification (LFQ) values, as well as iTop3 39 values (sum of the intensities of the three most intense peptides); for the latter, variance stabilization 40 was used for the peptide normalization, and missing peptide intensities, if at least 2 evidences exist in a group, were imputed by drawing values from a Gaussian distribution of width 0.3 centered at the sample distribution mean minus 1.8x the sample standard deviation. Imputation at protein level for both iTop3 and LFQ was performed if there were at least two measured intensities in at least one group of replicates; missing values in this case were drawn from a Gaussian distribution of width 0.3 centered at the sample distribution mean minus 2.5x the sample standard deviation. Differential expression tests were performed using empirical Bayes (moderated t-test) implemented in the R limma (RRID:SCR_010943) package 41. The Benjamini and Hochberg 42 method was further applied to correct for multiple testing.
Immumoblotting
Cells were lysed in RIPA buffer with protease inhibitors, resolved on 4-15% Mini-Protean TGX gels (BioRad, 456-1084), and transferred to nitrocellulose membranes using the iBlot2 system (Thermo Fisher, IB23001). Blots were blocked for 1 hour at room temperature and then probed with primary antibodies against METTL3 (Cell Signaling, 96391; RRID: AB_2800261), KIF5C (Abcam, ab193352), GAPDH (Millipore, AB2302; RRID: AB_10615768), IFIT3 (Santa Cruz, sc-393512; RRID:AB_2857847), STAT1 (Cell Signaling, 14994; RRID:AB_2737027), phospho-STAT1 (Cell Signaling, 7649; RRID:AB_10950970), or beta-actin (Abcam, ab6276; RRID: AB_2223210) overnight in 5% milk or BSA (all Cell Signaling antibodies) in TBST at 4 °C. After 4 washes, blots were incubated with IRDye secondary antibodies (LI-COR: 926-68075, RRID:AB_10974977; 926-32211, RRID:AB_621843; 926-32210, RRID:AB_621842) in 5% milk for one hour at room temperature. After 4 washes blots were visualized on a LI-COR Odyssey Infrared Imaging System.
m6A-Immunoprecipitation
Total RNA was isolated with TRIzol according to the manufacturer instructions, and PolyA+ mRNA was isolated using Dynabeads Oligo(dT)25 (Invitrogen, 61002) and genomic DNA was removed using the DNA-free kit (Ambion, AM1906). 3.5 μg of mRNA was fragmented (Invitrogen, AM8740), along with 4 pmol each of positive and negative control mRNAs (New England Biolabs, E1610S). 10% of each sample was saved as input and the remainder was brought up to 500 μl with IP buffer (50 mM Tris pH 7.4, 150 mM NaCl, 0.5% NP-40) and coupled to 2 μl of m6A antibody (Abcam, ab151230; RRID: AB_2753144) at 4 °C for 2 hours. Antibody-bound mRNA was precipitated with 50 μl of Protein AG beads (Thermo Scientific, 88803) at 4 °C for 1 hour. Beads were then washed five time in IP buffer, and RNA was eluted in RNA Binding Buffer and purified using clean up columns (Zymo, R1013). RNA was reverse transcribed using random primers and SuperScript IV Reverse Transcriptase (Invitrogen, 18090010). The input fraction was split into two in order to run in parallel a no reverse transcriptase control to assess for any potential amplification of contaminating genomic DNA.
qPCR
RNA was extracted directly from cells using the RNeasy Mini Kit with an on column DNAse treatment. RNA was reverse transcribed using the FIREScript RT cDNA kit and random primers (Solis Biodyne, 06-15-00200) according to the manufacturer’s instructions. Quantitative real-time PCR was performed on the ViiA 7 system (Applied Biosystems) using HOT FIREPol EvaGreen qPCR mix (Solis Biodyne, 08-24-00020) following manufacturer’s instruction. Primer sequences are listed in Supplementary Table 1. All quantitative real-time PCR assays were carried out using three technical replicates. Relative quantification of quantitative real-time PCR data used GAPDH, actin, and HMBS as housekeeping genes.
Interferon Stimulation
Cells were treated with 200 units/mL of recombinant interferon alpha (Abcam, ab48750) for between 4 and 24 hours before Western blot analysis.
RESULTS
Low METTL3 expression is associated with advanced prostate cancer
Given the ability of m6A to influence gene expression, we asked whether m6A affects PCa progression. To determine if changes m6A might contribute to prostate cancer in patients, we assessed if the expression of the methyltransferase m6A “writer” complex members (METTL3, METTL14, WTAP, VIRMA, ZC3H13 and RBM15), the m6A “eraser” ALKBH5, or the m6A “reader” proteins (YTHDF1, YTHDF2, and YTHDF3) is related to any clinical parameters in advanced PCa. In a recent precision oncology study, we reported on 430 men with castration resistant prostate cancer (CRPC)43. We subset these patients based on the relative expression levels of each member into low (z-score of less than −1) and high (z-score of greater than 1) groups. We then assessed whether these groups demonstrated low androgen receptor (AR) signaling or high neuroendocrine scores, both important measures for a lack of response to anti-androgen therapy44.
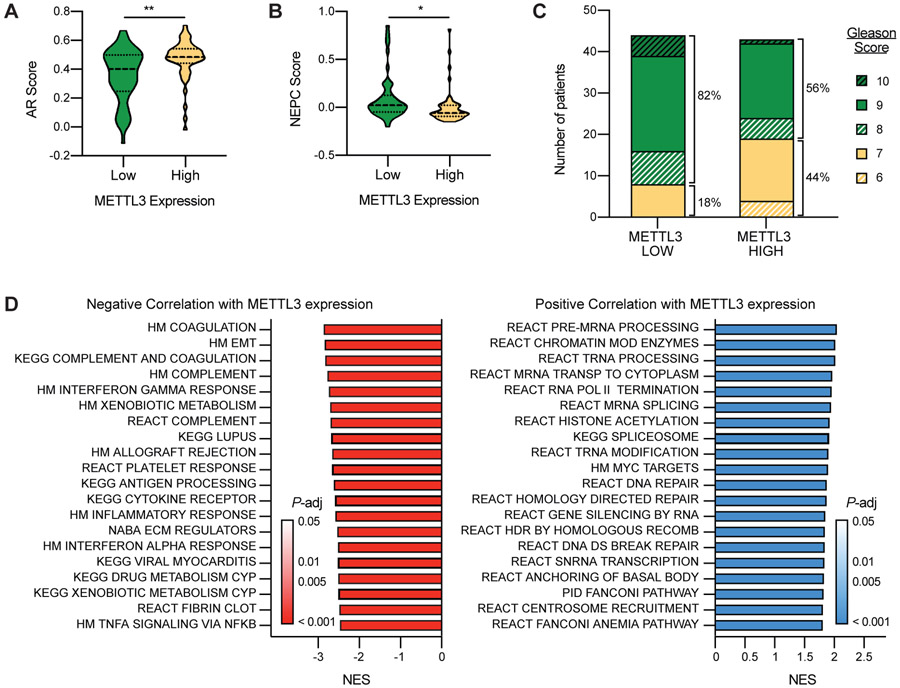
Although previous studies suggested that METTL3 expression is higher in primary PCa as compared to benign prostate tissue26,27, our examination of the expression of METTL3 in metastatic PCa samples revealed the opposite phenomena, that decreased METTL3 is associated with the most aggressive subclass of advanced prostate disease. In this cohort of 430 men with metastatic CRPC, we observed that those with lower levels of METTL3 demonstrated significantly disrupted AR signaling (P = 0.0011) (Figure 1A), and significantly increased expression of genes associated with neuroendocrine progression (P = 0.0214) (Figure 1B). This association between low expression and low AR score also held true for METTL14 (P = 0.0002), WTAP (P = 0.0002), VIRMA (P = 3.79E-06), ALKBH5 (P = 0.0034), YTHDF1 (P = 0.0323), YTHDF2 (P = 0.0024), and YTHDF3 (P = 0.0096) (Supplementary Figure 1A). In contrast ZC3H13 expression did not show a significant association with AR score (Supplementary Figure 1A), however it was the only other gene besides METTL3 to have a significant association with NEPC score, albeit in the opposite direction (P = 0.0433) (Supplementary Figure 1B).
Figure 1. Low METTL3 expression is associated with advanced-stage disease.
Castration resistant prostate cancer samples43 were grouped based on METTL3 expression: high, z-score > 1, n = 50; low, z-score < −1, n = 49. A) AR score in METTL3 high vs. low samples. Median and quartiles are indicated with a dotted line, P = 0.0011 as determined by t-test. B) NEPC score in METTL3 high vs. low samples. Median and quartiles are indicated with a dotted line, P = 0.0214 as determined by t-test. C) Gleason score in METTL3 high vs. low samples. P = 0.0111 as determined by Fischer’s exact test between samples with Gleason 6-7 and Gleason 8+ scores. D) Gene set enrichment analysis of genes differentially expressed in in METTL3 high vs. low samples. Genes were ranked according to the log of the Benjamini and Hochberg adjusted P-value as determined by t-test. Shown are the top 20 results in either direction.
In this same group of patients those with lower METTL3 expression were also more likely to have primary tumors with higher Gleason scores (8 and above) than those with high METTL3 expression (P = 0.0111) (Figure 1C). None of the other methyltransferase complex members, reader proteins, or ALKBH5 showed any association with Gleason score (Supplementary Figure 1C). Given these data, and its known role as the catalytic subunit of the “writer” complex we chose to focus further investigations on METTL3.
To begin to understand the mechanistic links between m6A and these signifiers of aggressive CRPC, we asked what genes and/or pathways are associated with differential METTL3 expression in PCa. To begin to address this question we took the same cohort of metastatic patient samples categorized into METTL3-high and METTL3-low groups, and looked for genes over- or under-expressed in one group versus the other. This analysis identified 2692 genes with significantly reduced expression in the METTL3-low group and 2081 genes with significantly increased expression. Gene set enrichment analysis of these genes demonstrates that those that correlate positively with METTL3 expression mainly involve genes associated with transcription and mRNA processing (Figure 1D). On the other hand, gene sets that display a negative correlation with METTL3 expression include many pathways implicated in aggressive metastatic disease, including complement and coagulation pathways, EMT, regulation of the extracellular matrix, and drug metabolism (Figure 1D).
Mapping m6A in prostate cancer
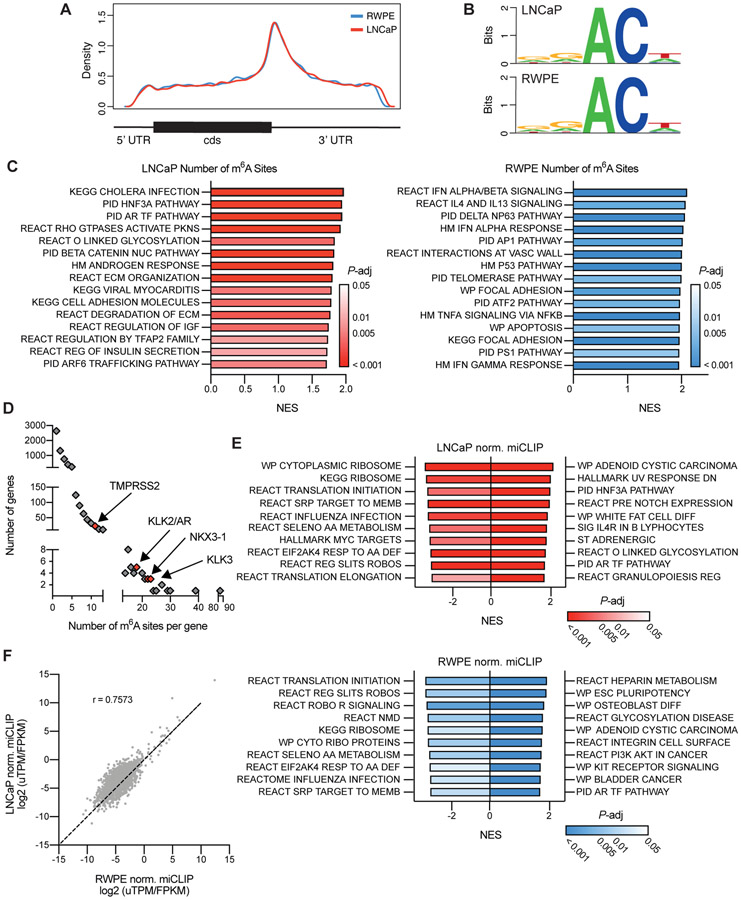
Given that our above analysis relies solely on correlations in gene expression within a diverse group of patient samples, we aimed to more thoroughly define the role of m6A methylation in the regulation of PCa gene expression. As a first step we sought to generate the first epitranscriptomic maps of m6A in PCa. Using m6A individual-nucleotide-resolution cross-linking and immunoprecipitation (miCLIP)31 we mapped the location of m6A at a single-nucleotide resolution throughout the entire transcriptomes of both the AR-sensitive adenocarcinoma LNCaP and in the benign RWPE cell line and identified over 18000 methylated residues in 6653 transcripts. Similar to previously reported m6A maps generated in other cell lines and tissues the majority of m6A sites identified in both cell lines were in the 3’ UTR, particularly in the vicinity of the stop codon (Figure 2A). Furthermore, the sequence context surrounding the methylation sites was also consistent with the previously reported DRACH motifs (D= A/G/U, R= A/G, H= A/C/U)14,45 (Figure 2B).
Figure 2. Mapping genes methylated with m6A in prostate cancer.
m6A was mapped in LNCaP and RWPE cell lines using miCLIP. A) Metagene plot of the distribution of m6A residues along the transcript in both cell lines. B) Consensus sequence surrounding m6A sites identified in both cell lines generated with WebLogo. C) Gene set enrichment analysis of genes ranked by the number of m6A sites. Shown are the top 15 gene sets. D) AR signature genes (n = 30; Hieronymus, 2006) are significantly (P = 0.0004) enriched in m6A sites (>10 sites) as determined by Fisher’s exact test. E) Gene set enrichment analysis of genes ranked by the total normalized miCLIP unique tags per million for a given transcript. F) Comparing the normalized miCLIP unique tags per million per transcript in genes which are similarly expressed (< 10-fold difference) between the LNCaP and RWPE call lines.
In both lines the 70-85% of transcripts had only one or two annotated sites, however 1-2% of the transcripts had 10 or more (Supplementary Data 1, 2). In both lines the non-coding RNA MALAT1 was the most highly methylated with 84 annotated sites in LNCaP and 69 in RWPE. Examination of the association between the total of number of annotated sites identified per transcript and its expression shows that while there is some bias towards highly expressed genes having more total sites (due to inherent biases in IP-based protocols), expression is not the sole driver of the differences seen (Supplementary Figure 2A). Furthermore, there was no correlation between the number of annotated m6A sites and the transcript length (Supplementary Figure 2B).
To determine the cellular pathways that m6A might influence in prostate cancer, we interrogated the miCLIP data using gene set enrichment analysis to identify pathways enriched in the transcripts that exhibited higher number of annotated m6A sites. This analysis implicated m6A in regulating mRNAs that encode pathways related to cell adhesion and the extracellular matrix in both cell lines (Figure 2C). Remarkably, this enrichment is similar to the gene expression changes linked to low METTL3 expression based on examination of the patient samples (see Figure 1D). In particular of the transcripts differentially expressed between the METTL3-high and -low groups 1430 and 684 exhibited at least one annotated m6A site in LNCaP or RWPE cells respectively (Supplementary Figure 2C). Additionally, this analysis identified AR-regulated genes as also having a high number of annotated m6A sites in LNCaP cells. In particular AR signature genes including KLK2, KLK3, NKX3-1 and even AR itself all had greater than 10 sites (Figure 2D).
We further characterized the methylation status of transcripts by not only the number of annotated sites on a given transcript, but also by a relative measurement of the amount of m6A at a given site. To do so we normalized the amount of fragmented RNA immunoprecipitated using the m6A antibody from each individual site to the library size (uTPM), and overall expression of that transcript (Supplementary Figure 2D). As in Supplementary Figure 2A, we see a correlation between the amount of immunoprecipitated RNA and the expression of the transcript. Nonetheless we can also identify distinct subsets of transcripts which have relatively high or low levels of methylation given their expression. In both cell lines this analysis again identified AR pathway genes as having high levels of methylation, whereas ribosomal and translation pathways had low levels of methylation relative to the expression levels of those transcripts (Figure 2E) in agreement with results from other human cell lines11.
Comparisons of methylation sites between the two lines is of course heavily biased by the highly divergent transcriptomes between LNCaP and RWPE cells. However, upon limiting the analysis to transcripts with less than a 10-fold difference in expression between the two lines we find a strong correlation between the amount of m6A (Pearson r = 0.7573) (Figure 2F). In particular, upon comparing the amount of methylation at m6A sites common to both cell lines we found a striking concordance between the levels in LNCaP and RWPE cells (Pearson r = 0.8666) (Supplementary Figure 2E).
METTL3 regulation of gene expression and translation
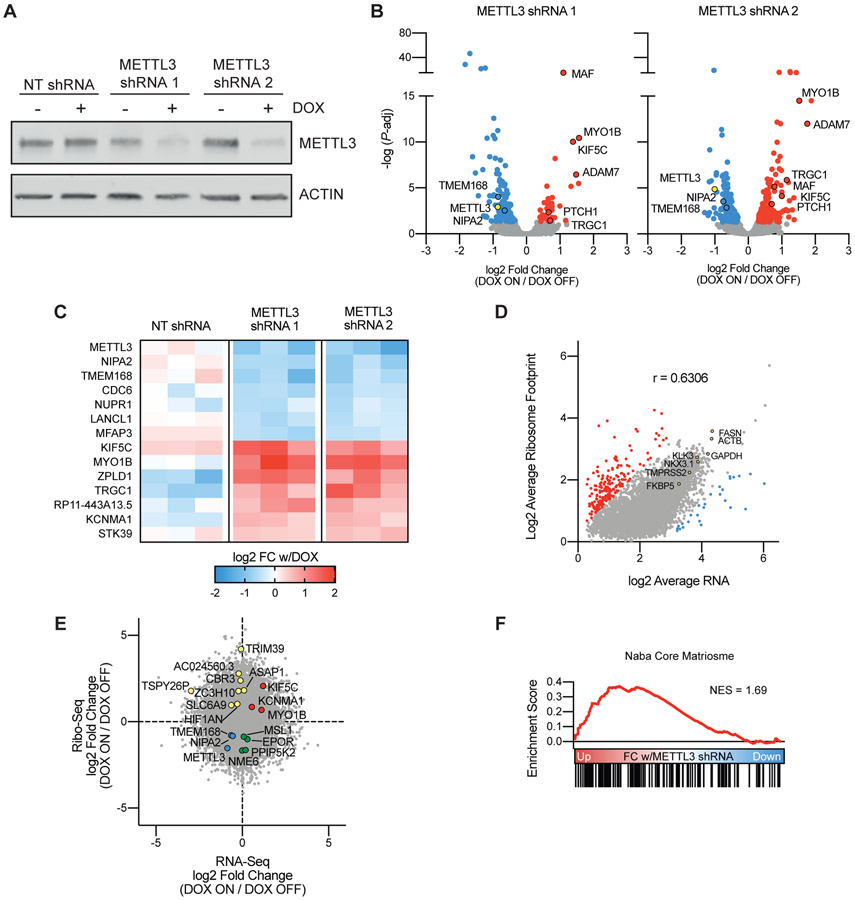
Although the miCLIP analysis identified many highly methylated transcripts with known functional relevance in PCa, in particular AR and its downstream target genes, it remains to be seen whether methylation of these transcripts plays a specific role in their regulation, and further whether this regulation is at the level of transcript stability and/or translation efficiency. To address this, we generated LNCaP cell lines infected with drug-inducible shRNAs targeting METTL3. With these lines we were able to precisely and coordinately manipulate both the timing and extent of reduction of METTL3 expression. We demonstrated a robust knockdown of METTL3 expression after 96 hours of doxycycline treatment (Figure 3A) as compared to a control non-targeting shRNA against GFP. Further the METTL3 knockdown is reversible after 144 hours of doxycycline wash-out (Supplementary Figure 3A).
Figure 3. Knockdown of METTL3 in prostate cancer alters gene expression and translation.
A) Generation of LNCaP cell lines with two doxycycline-inducible shRNAs targeting METTL3, or a non-targeting shRNA targeting GFP. Cells were treated with doxycycline for 96 hours followed by Western blot analysis for METTL3. B) Significantly (adjusted P-value < 0.05) differentially expressed genes with METTL3 knockdown in the two inducible shRNA lines (n = 3). Common genes between the two shRNA lines are highlighted with a black border. C) Six transcripts are significantly down-regulated and seven transcripts are significantly up-regulated with METTL3 knockdown in both shRNA lines, but not the non-targeting line (n = 3). D) Correlation (Pearson r = 0.6306, P < 1E-15) between the average mRNA (n = 6) and ribosome footprint (n = 4) from the METTL3 shRNA lines without doxycycline. Outliers with extreme high (red) and low (blue) translation efficiency (Ribo/RNA) were calculated using ROUT analysis (Q = 1%). E) Riborex analysis of Ribo-seq data identities genes with changes in TE independent of changes in mRNA expression. Shown is the fold change with doxycycline treatment in both METTL3 shRNAs combined as determined by DESeq2 (RNA-seq n = 12, Ribo-Seq n = 10). F) Gene set enrichment analysis of genes ranked by the fold change in TE with METTL3 knockdown as determined by Riborex.
Using these inducible lines, we measured changes in both expression and translation efficiency by means of tandem RNA-seq and ribosome footprint profiling (Ribo-Seq). RNA-seq analysis demonstrated relatively modest changes in gene expression with METTL3 knockdown. With either METTL3 shRNA we found 187 and 337 significantly (FDR < 0.05) differentially expressed genes between the doxycycline on and off conditions (Figure 3B). However, we also identified 508 significant gene expression changes in the non-targeting line in response to doxycycline, including three which were also significant in both our METTL3 shRNA lines (Supplementary Figure 3B). This strategy allowed us to specifically identify six down- and seven up-regulated transcripts (Figure 3C) which were significantly regulated in two different METTL3 knockdown lines, after considering non-specific effects of doxycycline treatment or shRNA induction. Of these thirteen transcripts, seven were identified in our previous miCLIP experiments to have at least one m6A mark in LNCaP cells (Supplementary Data 1).
To determine if these changes persisted at the protein level, we validated one of the most up-regulated transcripts KIF5C. We confirmed persistent KIF5C upregulation at the protein level after only 72 hours of doxycycline treatment (Supplementary Figure 3C). KIF5C is further of interest in that in metastatic PCa samples its expression is highly correlated with androgen receptor signaling (Spearman’s ρ = 0.56) (Supplementary Figure 3D). Given that expression of KIF5C itself is not regulated by androgen receptor signaling in LNCaP cells (Fold-change < 2 with DHT or ENZ) (Supplementary Figure 3E), it remains to be seen whether KIF5C operates upstream of AR, or if it plays some other role in PCa progression.
Similar to published Ribo-seq results from other cell lines and tissues46 ribosome footprinting read counts generally correlated with mRNA expression in LNCaP cells (Pearson r = 0.6306) (Figure 3D). Among the most highly expressed and ribosome-bound transcripts are the housekeeping genes GAPDH and ACTB, and the known AR-targets KLK3, FASN, NKX3.1, FKBP5, and TMPRSS2. We did, however, also identify 225 transcripts that were outliers (ROUT analysis, Q = 1%) with ribosome footprints that do not correlate with mRNA expression. 8 of the 27 transcripts with exceptionally low translation efficiency (TE) are mitochondrial genes which are known to be inefficiently captured with standard ribosome footprinting protocols47. Other gene sets with generally low TE include ribosomal proteins and pathways involved in translation (similar to results from proteogenomic studies demonstrating a low correlation between transcript and protein for ribosomal proteins in human tumors48-50), while those with high TE are predominated by extracellular matrix proteins in particular collagens (Supplementary Figure 3F).
We then utilized Riborex36 analysis to identify genes with significant changes in TE independent of any changes in mRNA expression. This analysis further identified 15 and 6 genes with a significant (FDR < 0.1) increase or decrease in TE with METTL3 knockdown in both of the inducible shRNA lines (Figure 3E), of which 6 were have at least one m6A mark in LNCaP cells (Supplementary Data 1). Gene set enrichment analysis demonstrated that the TE of transcripts encoding extracellular matrix proteins is increased with METTL3 knockdown (Figure 3F).
METTL3 regulation of protein expression
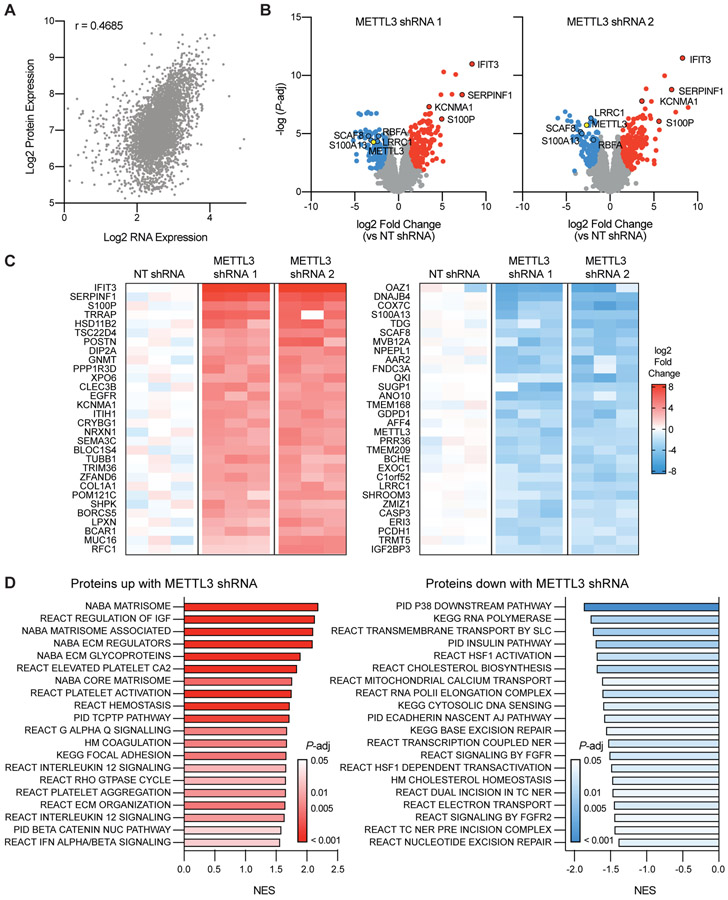
Given the changes we detected in translation efficiency with METTL3 knockdown we then wanted to understand how these changes are reflected in protein abundance. Therefore, we conducted shot-gun proteomics using the same inducible METTL3 shRNA lines in order to get an unbiased and quantitative picture of the entire proteome. Protein expression generally correlated with mRNA expression (Spearman’s ρ = 0.443-0.469) in line with recent results describing paired RNA-seq and proteomics in PCa patient samples6 (Figure 4A).
Figure 4. Knockdown of METTL3 in prostate cancer alters protein expression.
A) Correlation (Spearman’s ρ = 0.4685, P < 1E-15) between the average mRNA expression (n = 3) and protein expression (n = 3) as determined by shotgun proteomics in the NT shRNA line. B) Significantly (adjusted P-value < 0.05) differentially expressed proteins with METTL3 knockdown in the two inducible shRNA lines as compared to the NT shRNA line (n = 3 for each shRNA). Common highly differentially expressed proteins between the two shRNA lines are highlighted with a black border. C) The top 60 differentially expressed proteins seen with METTL3 knockdown in both shRNA lines as compared to the NT control (n = 3 for each shRNA). D) Gene set enrichment analysis of proteins differentially expressed with METTL3 knockdown. Proteins were ranked according to the log of the adjusted P-value as determined by t-test. Shown are the top 20 results in either direction.
We identified 527 and 537 significantly differentially expressed (FDR adj p-value < 0.05, FC > 2) proteins in either METTL3 shRNA line as compared to the non-targeting shRNA line (Figure 4B). Similar to our Ribo-seq results we also observed much larger fold changes at the protein level (> 15-fold) than we had seen at the level of expression (2-4-fold, Figure 3B). Unfortunately, only one of the transcripts with significant changes in TE were detected in our proteomics experiment, thus precluding a thorough comparison between the two methods; nonetheless this protein (CBR3) was upregulated in both cell lines after METTL3 knockdown. In total 182 proteins significantly upregulated in both METTL3 shRNA lines while 109 proteins were significantly down-regulated (Figure 4C, Supplementary Data 3), and of these 160 have at least one m6A mark (Supplementary Data 1). Previous studies have identified interferon signaling as being highly upregulated in response to METTL3 knockdown51,52. Here we identified an upregulation of the interferon-induced proteins IFIT1, IFIT2, and IFIT3, but not other interferon-induced proteins. This is likely due to the fact that LNCaP cells are unresponsive to interferon signaling (Supplementary Figure 4) due to a silencing of JAK1 by bi-allelic inactivating mutations and epigenetic silencing in this line53,54.
Gene set enrichment analysis of the proteomics data revealed that down-regulated proteins mainly included those associated with transcription and repair of DNA damage (Figure 4D). Gene sets with up-regulated proteins predominantly consisted of those involved in the composition and regulation of the extracellular matrix, coagulation and cell adhesion. This is consistent with results seen in patients with low METTL3 expression (Figure 1D) and genes with high levels of m6A in LNCaP cells (Figure 2D).
METTL3 knockdown triggers AR-independence
Recent work has demonstrated that the majority of the effects of m6A is mediated through a reduction in transcript stability16,55. Given this, one would expect that we would see an increase in expression of the highly methylated AR pathways genes (Figure 2D), and therefore AR signaling, with METTL3 knockdown. In contrast, we see no change in the expression of these genes at the transcript (Figure 3C) or protein level (Figure 4C). Furthermore, the expectation of an increase in AR signaling with METTL3 knockdown is inconsistent with the results we saw in the metastatic patient cohort where low levels of METTL3 were associated with a lower AR score, indicating a disruption of AR signaling overall (Figure 1A).
To resolve these contradictions, we decided to directly examine the response of the METTL3 knockdown lines to treatment with enzalutamide (ENZ, a potent AR ligand binding inhibitor), and/or stimulation with dihydroxytestosterone (DHT). We then examined the changes in expression of known AR target genes (i.e., KLK3, TMPRSS2, FKBP5 and NKX3.1) by qPCR (Figure 5A). In the absence of METTL3 knockdown all three cell lines responded similarly to the treatments. Further, with METTL3 knockdown we saw no change in the response of KLK3, FKBP5 and TMPRSS2 to AR inhibition with ENZ or charcoal-stripped (hormone-free) media, nor to DHT stimulation. In contrast, we did see a significant reduction in the down-regulation of NKX3.1 with METTL3 knockdown and ENZ (sh1, P = 0.0062; sh2, P = 0.0074) or charcoal-stripped media (sh1, P = 0.036; sh2, P = 0.044) treatment. NKX3.1 is a prostate-specific homeobox gene that regulates normal prostate differentiation and suppresses PCa initiation56. The upregulation of NKX3.1 with DHT stimulation, however, remained unchanged (FC sh1 = 2.3, FC sh2 = 2.6, vs FC NT = 3). As such, we concluded that METTL3 knockdown does not alter AR signaling overall in LNCaP cells.
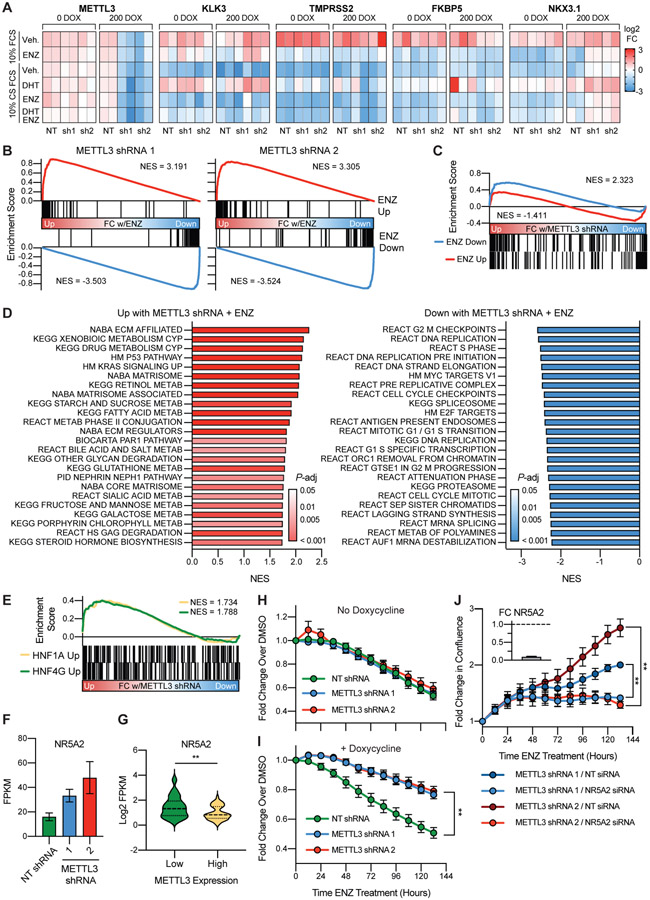
Figure 5. METTL3 knockdown leads to the induction of a gene signature linked to castration resistance and renders the cells resistant to enzalutamide in an androgen receptor independent manner.
A) Change in expression of AR target genes with METTL3 knockdown (+/− doxycycline) and in response to AR stimulation (10 nM DHT) or inhibition (10 μM ENZ). Expression was measured via qPCR (n = 2) and is displayed as the fold change with treatment per gene and experiment. B) RNA-seq analysis of METTL3 knockdown lines treated with ENZ demonstrates no change in the repression or activation of known ENZ target genes by gene set enrichment analysis. Genes were ranked according to the fold change ENZ vs vehicle as determined by DESeq2. ENZ up and down gene lists contain the common differentially expressed genes from two published RNA-seq experiments (GSE110903 and GSE147250). C) Some ENZ responsive genes show differential expression with METTL3 knockdown. Ranking for gene set enrichment analysis was according to the combined fold change ENZ vs vehicle as determined by DESeq2 for both METTL3 shRNA lines. D) Gene set enrichment analysis of genes differentially expressed with ENZ treatment and METTL3 knockdown. Genes were ranked according to the combined Wald statistic from DESeq2 for both METTL3 shRNA lines as compared to the NT shRNA. Shown are the top 24 results in either direction. E) METTL3 knockdown with ENZ treatment induces the expression of HNF1A and HNF4G target genes. Genes were ranked according to the combined Wald statistic from DESeq2 for both METTL3 shRNA lines as compared to the NT shRNA. HNF target genes were taken from previously published overexpression experiments in LNCaP cells (GSE85559). F) NR5A2 is overexpressed with METTL3 knockdown and ENZ treatment (n = 3, error bars = SEM) G) NR5A2 expression in METTL3 high vs. low samples (see Figure 1). Median and quartiles are indicated with a dotted line, P = 0.0034 as determined by t-test. H) In the absence of METTL3 knockdown the shRNA lines respond similarly to 10 μM ENZ treatment (n = 3, error bars = SEM). I) Induction of METTL3 knockdown renders the cells resistant to ENZ treatment (n = 4, error bars = SEM; sh1 P = 0.0012, sh2 P = 0.0011, as determined by t-test). For both panels shown is the change in cell confluency as compared to DMSO with time as measured via Incucyte imaging. J) Knockdown of NR5A2 significantly reverses the ability of the METTL3 knockdown cells to proliferate in the presence of 10 μM ENZ (n = 3, error bars = SEM; sh1 P = 0.001003, sh2 P = 0.001223, as determined by t-test). Plotted is the fold change in confluency compared to time zero. Sufficient NR5A2 knockdown (91%) was confirmed in tandem by qPCR and is plotted in the top left as fold change over NT siRNA (n = 3).
In order to get a global view of the response to METTL3 knockdown and AR inhibition with ENZ, we analyzed changes in gene expression with RNA-seq. As expected, we again confirmed that METTL3 knockdown did not change known AR signaling pathways, as most canonical ENZ responsive genes responded as expected to the treatment (Figure 5B). Nonetheless we did identify some genes, which similar to NKX3.1, demonstrated a reduction in the magnitude of their repression or activation with ENZ (Figure 5C). For example, MAML2 is down-regulated 5.8-fold with ENZ and the NT shRNA, yet only 3.48- and 2.92-fold with the METTL3 shRNAs (Supplementary Figure 5A).
Overall 54 genes were significantly up-regulated with both METTL3 shRNAs and under ENZ treatment conditions, while 25 genes were significantly down-regulated (FDR < 0.05, Supplementary Data 4). Of these 79 differentially expressed transcripts, 40 were identified as having at least one annotated m6A site in our miCLIP analysis of LNCaP cells (see Supplementary Data 1). Importantly the upregulated genes included many which have been previously linked to ENZ resistance including: TMEFF2, ADAMTS157, and COL5A258. Gene set enrichment analysis of genes which are differentially expressed with METTL3 knockdown in response to ENZ again identified an enrichment in pathways related extracellular matrix components (Figure 5D), which is consistent with our above results in patient expression data (Figure 1D), m6A mapping (Figure 2C), ribosome footprinting (Figure 3F), and proteomics (Figure 4D) analysis.
Additionally, we also identified many pathways involved in the metabolism of drugs and other compounds that were significantly upregulated with METTL3 knockdown, again consistent with expression results from metastatic patient samples (Figure 1D). These same pathways, in addition to pathways that include complement and coagulation factors (which we see upregulated with METTL3 knockdown Figures 1D/4D), make up a gastrointestinal-lineage transcriptional signature previously reported in PCa by Shukala, et al. This signature is regulated by HNF1A and HNF4G, and importantly leads to castration resistance59. Upon closer examination of our RNA-seq data, we specifically see a significant upregulation in genes which are upregulated by both HNF1A and HNF4G in LNCaP cells59 (Figure 5E). Although we do not see changes in the expression of either of these two transcription factors, we do see an upregulation of NR5A2 (Figure 5F), an important gastrointestinal transcription factor which has been shown to regulate the same pathways in breast and colon cancer cells60,61. Further, NR5A2 (also known as LRH-1) has also been previously linked to castration resistant prostate cancer, and in particular overexpression of NR5A2 in LNCaP cells rendered them resistant to the AR inhibitor bicalutamide, and to castration in a xenograft model62. Examination of the patient data presented in Figure 1, also revealed that there is an inverse correlation with NR5A2 and METTL3 expression in CRPC patient samples (Figure 5G).
Given these results we then chose to examine how the METTL3 knockdown lines respond at the cellular level to ENZ treatment, and if upregulation of NR5A2 and these gene signatures leads to a resistant phenotype in our model. In the absence of METTL3 knockdown all three cell lines responded similarly to 10 μM ENZ treatment, demonstrating an approximately 50% reduction in cell proliferation as compared to DMSO controls after 132 hours (Figure 5H). Strikingly, upon doxycycline-induced expression of the METTL3 shRNAs, both lines showed a significant (sh1, P = 0.0012; sh2, P = 0.0011) resistance to the ENZ treatment with only an 20% reduction in proliferation after 132 hours (Figure 5I).
In order to determine the mechanism behind this phenotype we combined our inducible knockdown of METTL3 with transient knockdown via siRNAs. Given that our RNA-seq analysis demonstrated normal transcriptional responses of known AR-target genes in response to ENZ (Figure 5B) we hypothesized that the mechanism of resistance to ENZ is independent of AR itself. This hypothesis was confirmed, in that combining knockdown of AR and METTL3 did not significantly change the ability of the cells to proliferate in the presence of ENZ (Supplementary Figure 5B). In contrast, knockdown of NR5A2 resulted in a significant reduction in the ability of the METTL3 knockdown lines to proliferate with ENZ treatment (Figure 5J). Taken together these findings suggest that knockdown of METTL3 promotes ENZ resistance through an AR-independent upregulation of a gene signature driven by NR5A2.
DISCUSSION
Current understanding of the effects of m6A is that the mark mostly serves to destabilize mRNAs16,55. As such, we would expect to see increases in the expression of methylated genes with METTL3 knockdown. Strikingly, in our data we see relatively few changes in mRNA expression with METTL3 knockdown. Furthermore, while changes in the translation of methylated transcripts are generally rarer, they predominantly serve to increase TE. Again, we would then expect to see a decrease in TE, whereas we see many increases at the level of TE and protein with METTL3 knockdown in PCa. It therefore remains likely that we are also identifying some secondary-level changes due to alterations in the expression of upstream m6A methylated genes. It should also be noted that overall, our expression changes are fewer and more modest than many published studies. We propose that this is likely due to the fact that we are engineering an ~70-80% knockdown instead of a complete METTL3 knockout. In support of these hypotheses, we attempted to determine whether METTL3 knockdown altered the levels of m6A on several candidate genes that were differentially regulated (MFAP3, BCHE, NIPA2, SPON2, TMEM168, and ZNF460) by m6A-IP followed by qPCR. While we confirmed m6A methylation at all the annotated sites we queried, and saw a trend towards decreased enrichment on several transcripts with METTL3 knockdown, they did not reach statistical significance (n = 4 independent experiments) (Supplementary Figure 6). It remains likely that the levels of knockdown we achieved elicit small but significant changes that are potentially below the sensitivity of this assay, or in a limited subset of genes beyond our candidates, but are sufficient to result in the observed phenotypic changes in these cells. While it may be possible to see stronger effects on gene expression with a METTL3 knockout this phenotype is not clinically relevant as there is no complete loss of METTL3 seen in PCa patients.
When examining pathways which are regulated by m6A in PCa, we consistently saw enrichment for extracellular matrix (ECM) proteins in both our cell line models (Figure 3F/4D/5D) and in CRPC patients (Figure 1D) with low levels of METTL3. Changes in the ECM are associated with increased tumor growth, migration and invasion, allowing for metastatic progression63,64, including in PCa65. Furthermore, changes in the ECM have been implicated in therapy resistance in breast, ovarian and PCa66-69. As such, it remains possible that these changes also contribute to the ENZ resistance demonstrated by the METTL3 knockdown cells, and this would be a focus of future studies, in particular in a 3-D cell culture environment.
Lastly, our analysis of CRPC patient data demonstrated an inverse correlation between the levels of METTL3 and AR signaling. When we examined the interplay between m6A and AR signaling we showed that though AR target genes and AR itself have relatively high numbers of annotated m6A sites, knockdown of METTL3 did not change expression of these genes at the transcript or protein level. This may be due to low stoichiometry of the m6A sites on these transcripts (unresolved by miCLIP technology), redundancy with other components of the methyltransferase complex, or a lack of functionality of m6A on these transcripts. Nonetheless, we did find that knockdown of METTL3 led to an upregulation of a previously identified gastrointestinal-lineage signature and rendered the cells resistant to ENZ in an AR-independent manner. Previously, this signature was shown to be driven by HNF4G and HNF1A which were induced in response to androgen deprivation59. Here we hypothesize that this signature is driven by an upregulation of another GI-lineage transcription factor NR5A2 in response to METTL3 knockdown. Although NR5A2 itself was not identified as a methylated transcript in our miCLIP analysis, it is worth noting that the basal expression of NR5A2 in LNCaP cells is relatively low. Therefore, it is possible that any potential methylation of this transcript may evade detection with an IP-based approach. Nonetheless it also plausible that METTL3 knockdown is altering the methylation a yet unidentified upstream regulator of NR5A2 as opposed to the gene itself. The exact mechanism behind how this signature leads to ENZ resistance remains to be determined. However, among the included genes are those involved in both the metabolism of steroid hormones (i.e. AKR1C3 and UGT2B15) or general drug metabolism (i.e. GSTA1, GSTA2, GSTK1) both of which would play a probable role in ENZ resistance.
In summary, in this study we identified many genes and pathways which are dysregulated in the context of low METTL3 expression in PCa. In particular we have nominated extracellular matrix proteins as being highly influenced by changes in METTL3 expression. Furthermore, we showed that combining METTL3 knockdown and ENZ treatment led to an AR-independent upregulation of a GI-specific gene signature driven by NR5A2 and rendered the cells resistant to ENZ. Future work will focus on delineating the precise mechanism behind the ENZ resistant phenotype in an attempt to determine whether NR5A2 and/or other downstream pathway genes may function as potential therapeutic targets in CRPC. Overall, these findings support a new role for m6A in regulating therapeutic sensitivity to ARSi, and furthermore suggests that patients with low levels of METTL3 expression may differentially respond to ARSi.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the members of the Rubin laboratory for their comments and suggestions. We thank the Weill Cornell Epigenomics Core and Viola Paradisio (Department of Biomedicine, University of Basel) for their assistance in running the high-throughput sequencing. We also thank the University of Bern Proteomics Core for their assistance with the proteomics experiments. We acknowledge expert assistance from Mariana Ricca at the University of Bern in preparing the manuscript for submission.
FINANCIAL SUPPORT
This project was supported by funding from the Prostate Cancer Foundation (18YOUN06, K.A.C.), the Weill Cornell Medicine SPORE in Prostate Cancer (Developmental Research Project, S.R.J. and M.A.R.), the Swiss Cancer League (KFS-4988-02-2020-R, S.P.), and the NIH (R01CA186702, S.R.J.).
Footnotes
COMPETING INTERESTS
S.R.J. is founder, advisor to, and owns equity in Gotham Therapeutics.
RESOURCE AVAILABILITY
Materials Availability
Inducible shRNA plasmids generated in this study have been deposited to Addgene (Plasmid #:162984, RRID:Addgene_162984; 162985, RRID:Addgene_162985; and 163017, RRID:Addgene_163017).
Cell lines generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
The miCLIP, RNA, and Ribo-Seq data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus 70 and are accessible through GEO Series accession number GSE161304 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE161304).
Raw images of blots in the paper have been deposited to Mendeley Data: http://dx.doi.org/10.17632/xv2vzc8pzz.1
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository71 with the dataset identifier PXD022348.
REFERENCES
- 1.Baca SC et al. Punctuated evolution of prostate cancer genomes. Cell 153, 666–677, doi: 10.1016/j.cell.2013.03.021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger MF et al. The genomic complexity of primary human prostate cancer. Nature 470, 214–220, doi: 10.1038/nature09744 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbieri CE et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet 44, 685–689, doi: 10.1038/ng.2279 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chalmers ZR et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 9, 34, doi: 10.1186/s13073-017-0424-2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varambally S et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell 8, 393–406, doi: 10.1016/j.ccr.2005.10.001 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Sinha A et al. The Proteogenomic Landscape of Curable Prostate Cancer. Cancer Cell 35, 414–427 e416, doi: 10.1016/j.ccell.2019.02.005 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desrosiers R, Friderici K & Rottman F Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A 71, 3971–3975 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bokar JA, Shambaugh ME, Polayes D, Matera AG & Rottman FM Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3, 1233–1247 (1997). [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10, 93–95, doi: 10.1038/nchembio.1432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ping XL et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res 24, 177–189, doi: 10.1038/cr.2014.3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartz S et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell Rep 8, 284–296, doi: 10.1016/j.celrep.2014.05.048 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol 16, 191–198, doi: 10.1038/ncb2902 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng G et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49, 18–29, doi: 10.1016/j.molcel.2012.10.015 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dominissini D et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206, doi: 10.1038/nature11112 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Wang X et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120, doi: 10.1038/nature12730 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaccara S & Jaffrey SR A Unified Model for the Function of YTHDF Proteins in Regulating m(6)A-Modified mRNA. Cell 181, 1582–1595 e1518, doi: 10.1016/j.cell.2020.05.012 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161, 1388–1399, doi: 10.1016/j.cell.2015.05.014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res 24, 1403–1419, doi: 10.1038/cr.2014.151 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alarcon CR, Lee H, Goodarzi H, Halberg N & Tavazoie SF N6-methyladenosine marks primary microRNAs for processing. Nature 519, 482–485, doi: 10.1038/nature14281 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui Q et al. m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep 18, 2622–2634, doi: 10.1016/j.celrep.2017.02.059 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang C et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A 113, E2047–2056, doi: 10.1073/pnas.1602883113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S et al. m6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 31, 591–606 e596, doi: 10.1016/j.ccell.2017.02.013 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin S, Choe J, Du P, Triboulet R & Gregory RI The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell 62, 335–345, doi: 10.1016/j.molcel.2016.03.021 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vu LP et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nature medicine 23, 1369–1376, doi: 10.1038/nm.4416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbieri I et al. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature 552, 126–131, doi: 10.1038/nature24678 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai J et al. RNA m(6)A Methyltransferase METTL3 Promotes The Growth Of Prostate Cancer By Regulating Hedgehog Pathway. Onco Targets Ther 12, 9143–9152, doi: 10.2147/OTT.S226796 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan Y, Du Y, Wang L & Liu X The M6A methyltransferase METTL3 promotes the development and progression of prostate carcinoma via mediating MYC methylation. J Cancer 11, 3588–3595, doi: 10.7150/jca.42338 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scher HI & Sawyers CL Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. J Clin Oncol 23, 8253–8261, doi: 10.1200/JCO.2005.03.4777 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Rubin MA, Bristow RG, Thienger PD, Dive C & Imielinski M Impact of Lineage Plasticity to and from a Neuroendocrine Phenotype on Progression and Response in Prostate and Lung Cancers. Mol Cell 80, P562–577, doi: 10.1016/j.molcel.2020.10.033 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roviello G et al. Role of the novel generation of androgen receptor pathway targeted agents in the management of castration-resistant prostate cancer: A literature based meta-analysis of randomized trials. Eur J Cancer 61, 111–121, doi: 10.1016/j.ejca.2016.04.002 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Linder B et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods 12, 767–772, doi: 10.1038/nmeth.3453 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olarerin-George AO & Jaffrey SR MetaPlotR: a Perl/R pipeline for plotting metagenes of nucleotide modifications and other transcriptomic sites. Bioinformatics 33, 1563–1564, doi: 10.1093/bioinformatics/btx002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, doi: 10.1093/bioinformatics/bts635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li B & Dewey CN RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323, doi: 10.1186/1471-2105-12-323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li W, Wang W, Uren PJ, Penalva LOF & Smith AD Riborex: fast and flexible identification of differential translation from Ribo-seq data. Bioinformatics 33, 1735–1737, doi: 10.1093/bioinformatics/btx047 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cox J & Mann M MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology 26, 1367–1372, doi: 10.1038/nbt.1511 (2008). [DOI] [PubMed] [Google Scholar]
- 38.UniProt C UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47, D506–D515, doi: 10.1093/nar/gky1049 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silva JC, Gorenstein MV, Li GZ, Vissers JP & Geromanos SJ Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Molecular & cellular proteomics : MCP 5, 144–156, doi: 10.1074/mcp.M500230-MCP200 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Huber W, von Heydebreck A, Sultmann H, Poustka A & Vingron M Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18 Suppl 1, S96–104, doi: 10.1093/bioinformatics/18.suppl_1.s96 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Kammers K, Cole RN, Tiengwe C & Ruczinski I Detecting Significant Changes in Protein Abundance. EuPA Open Proteom 7, 11–19, doi: 10.1016/j.euprot.2015.02.002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benjamini Y & Hochberg Y Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B (Methodological) 57, 289–300, doi: 10.1111/j.2517-6161.1995.tb02031.x (1995). [DOI] [Google Scholar]
- 43.Abida W et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A 116, 11428–11436, doi: 10.1073/pnas.1902651116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beltran H et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nature medicine 22, 298–305, doi: 10.1038/nm.4045 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyer KD et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 149, 1635–1646, doi: 10.1016/j.cell.2012.05.003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ingolia NT, Ghaemmaghami S, Newman JR & Weissman JS Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223, doi: 10.1126/science.1168978 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rooijers K, Loayza-Puch F, Nijtmans LG & Agami R Ribosome profiling reveals features of normal and disease-associated mitochondrial translation. Nat Commun 4, 2886, doi: 10.1038/ncomms3886 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McDermott JE et al. Proteogenomic Characterization of Ovarian HGSC Implicates Mitotic Kinases, Replication Stress in Observed Chromosomal Instability. Cell Rep Med 1, doi: 10.1016/j.xcrm.2020.100004 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mertins P et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534, 55–62, doi: 10.1038/nature18003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang B et al. Proteogenomic characterization of human colon and rectal cancer. Nature 513, 382–387, doi: 10.1038/nature13438 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rubio RM, Depledge DP, Bianco C, Thompson L & Mohr I RNA m(6) A modification enzymes shape innate responses to DNA by regulating interferon beta. Genes Dev 32, 1472–1484, doi: 10.1101/gad.319475.118 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winkler R et al. m(6)A modification controls the innate immune response to infection by targeting type I interferons. Nat Immunol 20, 173–182, doi: 10.1038/s41590-018-0275-z (2019). [DOI] [PubMed] [Google Scholar]
- 53.Danziger O, Shai B, Sabo Y, Bacharach E & Ehrlich M Combined genetic and epigenetic interferences with interferon signaling expose prostate cancer cells to viral infection. Oncotarget 7, 52115–52134, doi: 10.18632/oncotarget.10313 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dunn GP, Sheehan KC, Old LJ & Schreiber RD IFN unresponsiveness in LNCaP cells due to the lack of JAK1 gene expression. Cancer Res 65, 3447–3453, doi: 10.1158/0008-5472.CAN-04-4316 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Lasman L et al. Context-dependent functional compensation between Ythdf m(6)A reader proteins. Genes Dev 34, 1373–1391, doi: 10.1101/gad.340695.120 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen MM & Abate-Shen C Roles of the Nkx3.1 homeobox gene in prostate organogenesis and carcinogenesis. Dev Dyn 228, 767–778, doi: 10.1002/dvdy.10397 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Kregel S et al. Acquired resistance to the second-generation androgen receptor antagonist enzalutamide in castration-resistant prostate cancer. Oncotarget 7, 26259–26274, doi: 10.18632/oncotarget.8456 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kohrt SE et al. Identification of genes required for enzalutamide resistance in castration-resistant prostate cancer cells in vitro. bioRxiv, 2020.2003.2027.011825, doi: 10.1101/2020.03.27.011825 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shukla S et al. Aberrant Activation of a Gastrointestinal Transcriptional Circuit in Prostate Cancer Mediates Castration Resistance. Cancer Cell 32, 792–806 e797, doi: 10.1016/j.ccell.2017.10.008 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bayrer JR, Mukkamala S, Sablin EP, Webb P & Fletterick RJ Silencing LRH-1 in colon cancer cell lines impairs proliferation and alters gene expression programs. Proc Natl Acad Sci U S A 112, 2467–2472, doi: 10.1073/pnas.1500978112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bianco S, Brunelle M, Jangal M, Magnani L & Gevry N LRH-1 governs vital transcriptional programs in endocrine-sensitive and -resistant breast cancer cells. Cancer Res 74, 2015–2025, doi: 10.1158/0008-5472.CAN-13-2351 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Xiao L et al. Nuclear Receptor LRH-1 Functions to Promote Castration-Resistant Growth of Prostate Cancer via Its Promotion of Intratumoral Androgen Biosynthesis. Cancer Res 78, 2205–2218, doi: 10.1158/0008-5472.CAN-17-2341 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Kai F, Drain AP & Weaver VM The Extracellular Matrix Modulates the Metastatic Journey. Developmental cell 49, 332–346, doi: 10.1016/j.devcel.2019.03.026 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Winkler J, Abisoye-Ogunniyan A, Metcalf KJ & Werb Z Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat Commun 11, 5120, doi: 10.1038/s41467-020-18794-x (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Penet MF et al. Structure and Function of a Prostate Cancer Dissemination-Permissive Extracellular Matrix. Clin Cancer Res 23, 2245–2254, doi: 10.1158/1078-0432.CCR-16-1516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hanker AB et al. Extracellular Matrix/Integrin Signaling Promotes Resistance to Combined Inhibition of HER2 and PI3K in HER2(+) Breast Cancer. Cancer Res 77, 3280–3292, doi: 10.1158/0008-5472.CAN-16-2808 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Januchowski R, Zawierucha P, Rucinski M, Nowicki M & Zabel M Extracellular matrix proteins expression profiling in chemoresistant variants of the A2780 ovarian cancer cell line. Biomed Res Int 2014, 365867, doi: 10.1155/2014/365867 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Joyce MH et al. Phenotypic Basis for Matrix Stiffness-Dependent Chemoresistance of Breast Cancer Cells to Doxorubicin. Front Oncol 8, 337, doi: 10.3389/fonc.2018.00337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mosquera MJ et al. Extracellular Microenvironment in Patient-derived Hydrogel Organoids of Prostate Cancer Regulates Therapeutic Response. medRxiv, 2020.2005.2017.20104349, doi: 10.1101/2020.05.17.20104349 (2020). [DOI] [Google Scholar]
- 70.Edgar R, Domrachev M & Lash AE Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30, 207–210, doi: 10.1093/nar/30.1.207 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perez-Riverol Y et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47, D442–D450, doi: 10.1093/nar/gky1106 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.