Fig. 1.
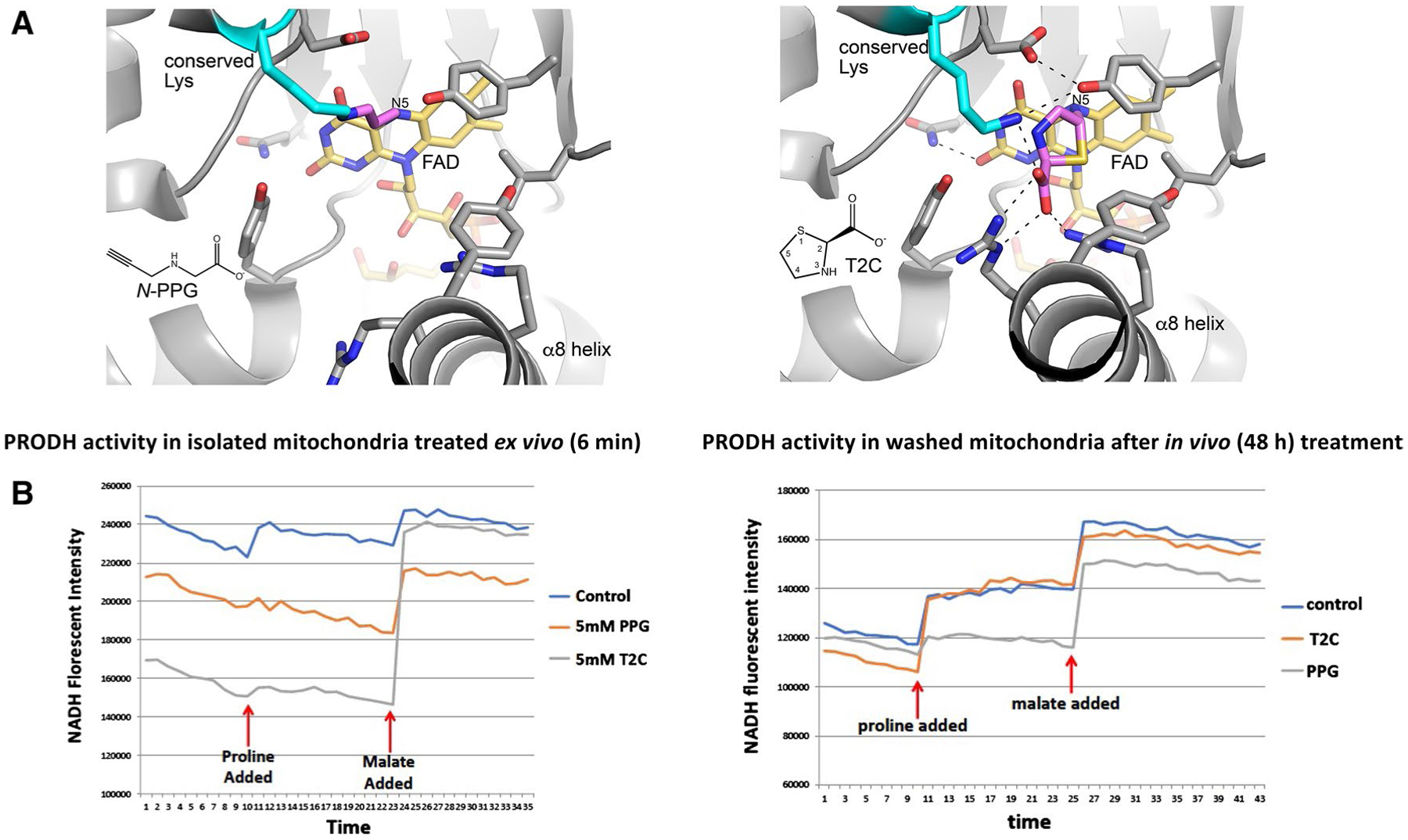
Structural models and enzymatic assays of PRODH inhibition by either N-PPG or T2C. A Structural basis for inactivation of PRODH by N-PPG (left panel) and the proline analog T2C (right panel). The left panel shows a crystal structure of a bacterial PRODH inactivated by N-PPG (Protein Data Bank ID 4NME). All the side chains shown are identically present in human PRODH. The conserved lysine (Lys234 in human PRODH) is colored cyan, the FAD is colored gold, and the 3-carbon covalent link is pink. The right panel shows the crystal structure of a bacterial PRODH inactivated by T2C (Protein Data Bank ID 6VZ9). All the side chains shown are identically present in human PRODH. The conserved lysine (Lys234 in human PRODH) is colored cyan, the FAD is colored gold, and covalently bound T2C is pink. B Treating isolated ZR-75-1 mitochondria with either N-PPG or T2C inhibits proline oxidation (left panel; x-axis units: 0.48 min/tick, total time shown = 17 min). However, isolating and then washing (15 min) mitochondria from control, N-PPG, or T2C pretreated (5 mM × 48 h) ZR-75-1 cell cultures and then assaying for NADH formation in the presence of rotenone by sequential addition of proline (1 mM) followed by malate (1 mM) shows full restoration of PRODH activity to control levels in T2C treated cells but persistent inhibition of proline oxidation and NADH formation in N-PPG pretreated cells (right panel; x-axis units: 0.48 min/tick, total time shown = 21 min)
