Fig 4. 4360 TCRs recognized RAS hotspot mutation: Validation, avidity and reactivity.
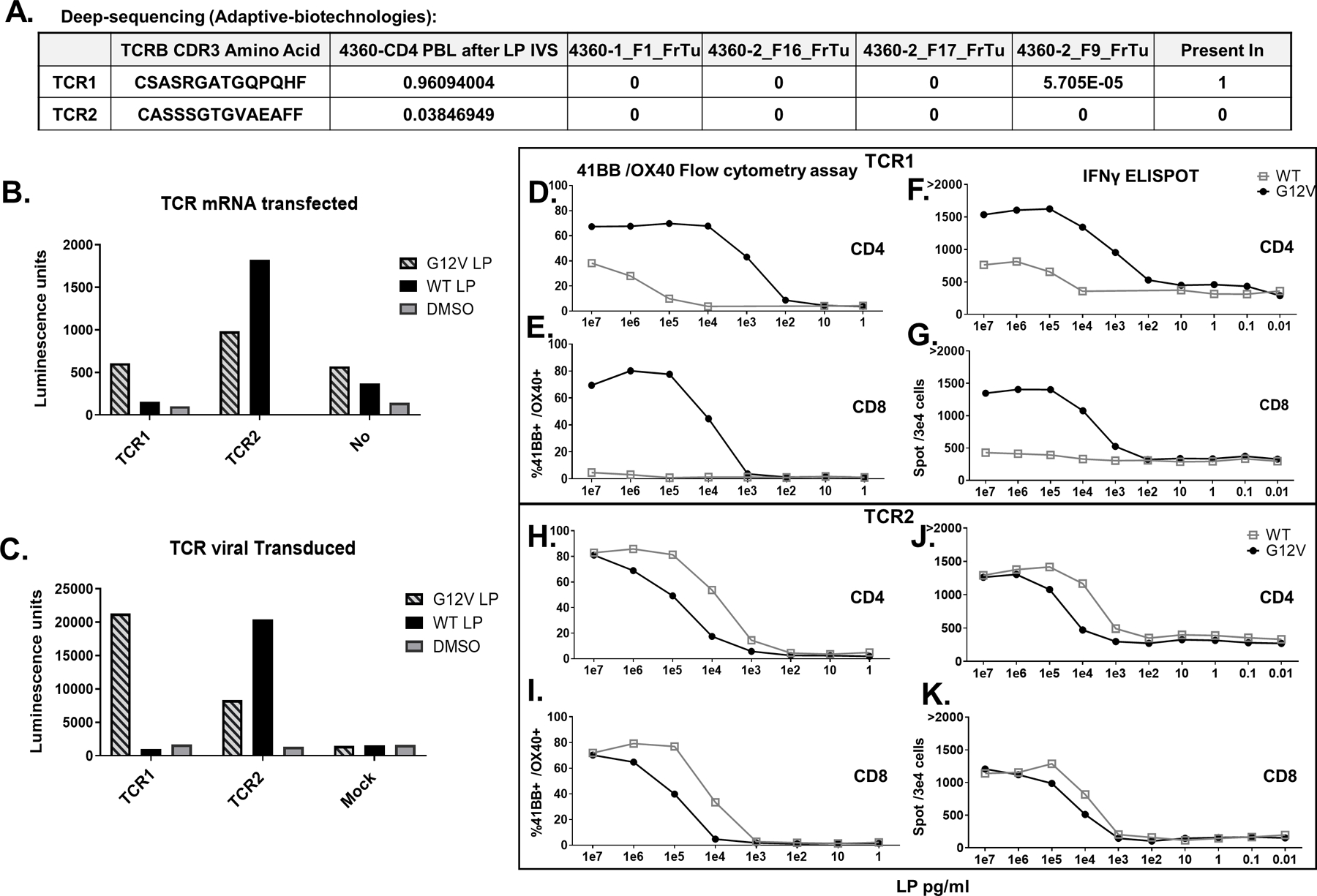
A-K. Two TCRs (TCR1, TCR2) identified from patient 4360 CD4 PBL after LP IVS single-cell sequencing. A. Deep sequencing (Adaptive Biotechnologies) of DNA extracts from four tumor fragments (FrTu) revealed TCR1 existed in one of these fragments (5.7 repeats in 100,000 cells) while TCR2 did not exist in any. B. Both TCRs were mRNA transfected or C. virally transduced into a Jurkat-CD4-NFAT-Luciferase cell line and then cocultured with DC loaded with RASG12V/ RASWT LP or the equivalent amount of DMSO. Luciferase activity was measured and presented in Luminescence units. D-G. TCR1 and H-K. TCR2 were virally transduced into patient 4360 PBLs. The cells enriched to CD3+/CD4+ cells D, F, H, and J or enriched to CD3+/CD8+ E,G,I, and K. The transduced cells were cocultured with DC loaded with different concentration of RASG12V/ RASWT LP. TCR avidity in live/CD3+/mTCR+ cells was analyzed by D, E, H, I. flow cytometry for 41BB/OX40 surface marker upregulation and F, G, J, K. IFN-γ ELISpot.
