Abstract
Conditional genetically engineered mouse models (GEMMs) of non-small cell lung cancer (NSCLC) harbor common oncogenic driver mutations of the disease, but in contrast to human NSCLC these models possess low tumor mutational burden (TMB). As a result, these models often lack tumor antigens that can elicit host adaptive immune responses, which limits their utility in immunotherapy studies. Here, we establish Kras-mutant murine models of NSCLC bearing the common driver mutations associated with the disease and increased TMB, by in vitro exposure of cell lines derived from GEMMs of NSCLC [KrasG12D (K), KrasG12DTp53−/−(KP), KrasG12DTp53+/−Lkb1−/− (KPL)] to the alkylating agent N-methyl-N-nitrosourea (MNU). Increasing the TMB enhanced host anti-tumor T cell responses and improved anti-PD-1 efficacy in syngeneic models across all genetic backgrounds. However, limited anti-PD-1 efficacy was observed in the KPL cell lines with increased TMB, which possessed a distinct immunosuppressed tumor microenvironment (TME) primarily composed of granulocytic myeloid-derived suppressor cells (G-MDSCs). This KPL phenotype is consistent with findings in human KRAS-mutant NSCLC where LKB1 loss is a driver of primary resistance to PD-1 blockade. In summary, these novel Kras-mutant NSCLC murine models with known driver mutations and increased TMB have distinct TMEs and recapitulate the therapeutic vulnerabilities of human NSCLC. We anticipate that these immunogenic models will facilitate the development of innovative immunotherapies in NSCLC.
Keywords: Mouse cancer models, NSCLC, TMB, KRAS, LKB1, Immunotherapy
Introduction
Immune checkpoint inhibitors (ICIs) targeting the PD-1/PD-L1 axis have resulted in durable clinical responses and improved survival in NSCLC [1]. However, most patients either do not respond to treatment or develop resistance to therapy after an initial response. Favorable responses to ICIs are associated with high TMB, preexisting intratumoral CD8+ T cells, and high baseline PD-L1 expression within the TME [2–4]. An increased number of candidate MHC class I tumor-neoantigens and a clonal neoantigen burden have also been associated with improved responses to ICIs in NSCLC [5, 6]. These results support the hypothesis that the non-synonymous somatic mutations associated with increased TMB generate tumor-neoantigens which can be recognized by host T cells as non-self. Treatment with ICIs can stimulate neoantigen-specific T cells to mediate tumor regression.
While ICIs have transformed the treatment landscape of NSCLC, a key impediment to progress in the field of lung cancer immunotherapy is the lack of preclinical models that recapitulate the complexity of human NSCLC. Human NSCLCs have among the highest mutational burden of all malignancies, and commonly possess driver mutations that confer distinct phenotypes [7]. KRAS mutations are the most prevalent oncogenic drivers in NSCLC and frequently co-occur with mutations in TP53 and LKB1, which define subgroups of patients with distinct biology and therapeutic vulnerabilities [8]. Although conditional GEMMs of Kras-mutant NSCLC possess the common driver mutations, studies reveal that these models harbor low TMB with few protein-altering mutations [9–11]. As a result, these GEMMs of NSCLC have limited value in the evaluation of host anti-tumor immune responses in translational preclinical immunotherapy studies [12]. To our knowledge, there are no reported murine models that possess both the critical driver mutations and relevant TMB observed in human NSCLC.
Herein, we report novel immunogenic Kras-mutant murine models of NSCLC bearing the common oncogenic mutations of the disease and increased TMB. These models possess unique TMEs and recapitulate the therapeutic vulnerabilities observed in human disease. Specifically, the cell line with LKB1-deficiency and increased TMB showed limited responses to anti-PD-1 therapy and therefore represents a clinically relevant preclinical model of NSCLC that is resistant to ICIs.
Materials and methods
Murine cell lines
Murine cell lines 1969B (labeled as the ‘KP’ cell line in the remainder of the manuscript), KP-2042, KP-1.1, and KP-3.1 were established from lung adenocarcinomas of conditional cre-lox-cre KrasG12DTp53−/−Luc FVB mice that express firefly luciferase (Supplemental Figure S1A). Murine cell lines 1940A (labeled as the ‘KPL’ cell line in the remainder of the manuscript), KPL-1950A, KPL-1942B, and KPL-2 were established from KrasG12DTp53+/−Lkb1−/−Luc (KPL) FVB mice. Whole exome sequencing (WES) analysis revealed that KPL cells lost the other allele of Tp53 upon in vitro culture and, therefore, bear a KrasG12DTp53−/−Lkb1−/−Luc genotype. The KrasG12D LKR-13 line (labeled as the ‘K’ cell line in the remainder of the manuscript), established from a lung adenocarcinoma tumor from a K-rasLA1 mouse, was generously provided by Dr. Jonathan Kurie [13]. Each cell line was maintained in culture media (RPMI-1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin) at 37 °C in a humidified atmosphere of 5% CO2.
In vivo studies
FVB and 129-E mice were purchased from Charles River Laboratories. Tumor cells were implanted in 7–9-week-old mice subcutaneously at optimal doses as indicated in figure legends. Tumor length and width were measured by caliper and the volume calculated by the equation: 0.4 × length × width2. For bioluminescence studies, images were obtained with the IVIS Spectrum imager after intraperitoneal (IP) injection of D-luciferin (150 mg/kg). For immunotherapy studies, mice bearing ∼ 50 mm3 tumors were randomized and treated with 200 μg of anti-PD-1 antibody (BioXcell, Clone RMP1–14) or isotype control via IP injections three times weekly for four doses. Mice were housed in pathogen-free facilities at UCLA and all procedures were approved by the UCLA Animal Research Committee.
Chemical treatment
Cells were seeded in T25 flasks and when ∼ 70% confluency was achieved, culture media was removed and the cells exposed to 100 μg/mL of MNU (Chem Service, NG-17031) in PBS for 45 min. After the removal of MNU, cells were washed with PBS twice and fresh culture media was added. Cells were passaged a minimum of three times prior to the subsequent MNU exposure for up to seven cycles.
In vitro proliferation assay
Cells were plated in culture media in 96-well plates at 1000 cells per well in eight replicates. Proliferation was measured using ATPlite 1step Luminescence Assay Kit (Perkin Elmer) every 24 h up to 120 h. Reading at each time point was normalized to the reading at baseline to control for plating differences.
In vitro IFN-γ stimulation
Cells were seeded in 6-well plates and treated with IFN-γ at 100 ng/mL when 50% confluency was achieved. Cells were harvested 24 h after stimulation and PD-L1 expression was analyzed by flow cytometry (FACS).
Tissue preparation
Spleens were mashed with the blunt end of 3 mL syringe on Petri dishes containing 5 mL of PBS, filtered through 70 μM filter, and centrifuged at 1500 rpm for 5 min at 4 °C. Cell pellets were resuspended in 5 mL of red blood cell lysis solution (BioLegend) on ice for 5 min followed by the addition of 20 mL of complete media. Cells were filtered through 70 μM filter, centrifuged, washed with PBS, and counted. Murine tumors were harvested, minced with scalpel blades, and digested in 2.5 mL of culture media containing 1 mg/mL of Collagenase IV (Roche) and 50 unit/mL DNase (Sigma) in 15 mL tubes at 37 °C with shaking every 10 min. After 45 min of incubation, 10 mL of fresh culture media was added and the samples were filtered through 70 μM filter and centrifuged. Red blood cells were lysed as described above, and the cells were washed with PBS, and counted.
FACS
Single-cell suspension from tumor, spleen, or cell culture were incubated with antibodies for 20 min at 4 °C followed by washing with staining buffer (PBS + 2% FBS). Intracellular staining was performed using an eBioscience intracellular kit according to the manufacturer’s protocol. FACS was performed on Attune NxT cytometer (ThermoFisher), and data analyzed by FlowJo software (TreeStar). Details of the flow antibodies utilized are listed in the Supplemental Table S1.
Genomic profiling
Genomic DNA isolation, library preparation, and sequencing
Genomic DNA was extracted from tumor cells (Qiagen, DNeasy blood and tissue kit) for WES. Tail DNA from three FVB and three 129-E mice was included as a normal reference for variant calls. Libraries for WES were prepared using the KAPA Hyper Prep Kit (Roche, KK8504) followed by exome enrichment with SeqCap EZ Share Developer Probe (Roche, 08333025001). Sequencing was performed on Hiseq3000 instrument as 150 bp pair-end runs with the aim of 100 × depth at UCLA TCGB Core facility.
Sequencing alignment
Sequence reads were aligned to the mouse genome (mm10) with Burrows-Wheeler Aligner (v 0.7.17), then marked for duplicates and re-calibrated as suggested by Genome Analysis Toolkit (GATK).
Variant calling and annotation
Strelka2 was utilized to call variants between cell line and the associated normal tail genome. When more than one reference normal genome was available (e.g. FVB mice), variant calls were performed to the individual normal genome independently and kept for down-stream analyses if they were called based on both reference genomes. Finally, a variant was called a mutation if (1) it was not in the germline mutation panel, determined from the FVB and 129-E tail genomes, (2) it was not supported by any read in the associated normal genome, (3) it was detected by at least 5 reads in cell lines, and (4) its variant allelic frequency (VAF) was > 0.1. The variants that passed these criteria were then annotated by Ensembl Variant Effect Predictor as non-synonymous mutations. Mutation with VAF ≥ 0.4 in all related cell lines including the parental cell line were defined as truncal. Shared mutations among members of the family with VAF < 0.4 were named as branch. Mutations with VAF < 0.4 which were specific to one cell line and not shared with the related cell lines were defined as private.
Neoantigen prediction
Mutations altering amino acid sequences of the encoded proteins were subjected to MHC-I binding prediction utilizing NetH2pan [14]. Peptides subjected to the algorithm were composed of 8–11 amino acid residues with the mutated amino acid at different locations. A candidate neoantigen had its affinity score ranked < 2nd percentile based on the binding software recommendation.
Statistical analysis
Statistical analyses were performed by GraphPad Prism version 7.04 for Windows. In brief, statistical significance was based on two-tailed non-paired Student’s t test for pairwise comparison and two-way ANOVA with Tukey post-test for time-associated comparison among multiple groups. Numerical data was presented as mean ± SEM.
Results and discussions
Establishing mouse models of NSCLC with known driver mutations and varying TMB
Given the low mutational burden in Kras-mutant GEMMs of NSCLC, we reasoned these tumors would be resistant to anti-PD-1 therapy. We screened cell lines established from Kras-mutant GEMMs of NSCLC with common co-occurring genomic alterations, namely K, KP, and KPL, with anti-PD-1 antibody in immunocompetent mice. Primary resistance to anti-PD-1 immunotherapy was observed across all cell lines, except for K, which showed a statistically significant but modest response to PD-1 blockade (Fig. 1a, and Supplemental Figures S2A, S2B and S2C). Because loss of a functional PD-L1 axis has been implicated in primary resistance to ICIs, we assessed the capacity of IFN-γ to upregulate PD-L1 in K, KP, and KPL cells in vitro [15]. IFN-γ stimulation resulted in the upregulation of PD-L1 in all cell lines examined, confirming an intact PD-L1 axis (Supplemental Figure S2D).
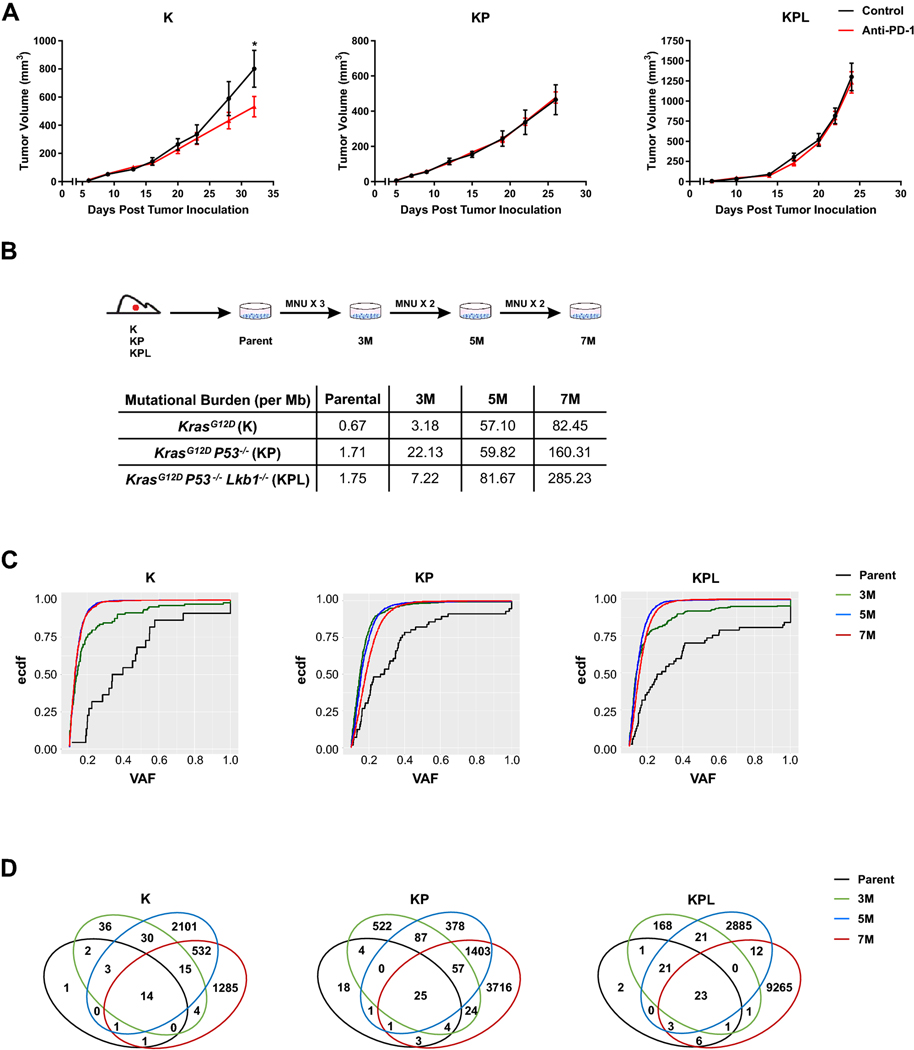
Fig. 1.
Murine models of NSCLC with varying mutational burden. a After subcutaneous (SC) tumor inoculation [K (2 × 106) cells in 129-E mice; KP (8 × 105) cells in FVB mice; KPL (1 × 105) cells in FVB mice], mice bearing < 50mm3 tumors (∼ day 7) were treated with (i) isotype control, (ii) anti-PD-1 (200 μg/dose 3 times weekly for 4 doses), and tumor growth was measured with caliper. Results are representatives of at least two biological replicates of 6–10 mice per group. b K, KP, and KPL were exposed to 100 μg/mL of MNU for 45 min. Cells were passaged prior to additional exposures to MNU for a total of 3, 5, and 7 exposures (3M, 5M, 7M). TMBs revealed by WES analyses are shown in the table. c Empirical cumulative distribution function (ECDF) of the mutations is plotted against VAF as an illustration of tumor heterogeneity within each family of cells. d Venn diagram of shared and private mutations of the K, KP, and KPL isogenic cell lines. P values were determined by two-way ANOVA with Tukey post-test. *P < 0.05
To establish Kras-mutant NSCLC cell lines that harbor the essential genomic drivers of the disease and increased TMB, we subjected one cell line from each genomic background, namely K, KP and KPL, to the carcinogen MNU for various durations (3, 5, and 7 exposures to MNU for 45 min each time, designated as 3M, 5M, and 7M, respectively) (Fig. 1b). Although we evaluated other tobacco related carcinogens for this purpose, namely benzyo[a]pyrene and nicotine-derived nitrosamine ketone (NNK), we found MNU to have the highest in vitro efficacy to increase the mutational load of cell lines as determined by WES. MNU is a widely utilized in vivo carcinogen to generate murine models of NSCLCs [9]. Although the mechanism of MNU-mediated DNA damage is distinct from tobacco smoking, its potency and wide utility, documented in the literature, suggests MNU as an effective agent for generating passenger mutations in NSCLC cell lines that already possess the critical driver mutations of the Kras-mutant NSCLC. WES revealed a dose-dependent increase in the number of non-synonymous mutations across all genomic backgrounds, resulting in higher TMBs (Fig. 1b). In parallel to increased TMB within each family of isogeneic cell lines, we observed an increased proportion of mutations with low VAF, indicating increased tumor heterogeneity (Fig. 1c). MNU did not result in changes in the genomic copy number of parental cell lines which were diploid. Evaluation of the copy number and mutations of the driver genes confirmed the deletion of Tp53 in KP cell lines and Tp53 and Lkb1 in KPL cell lines. Wild-type Tp53 was preserved in K cell lines. No additional Kras mutated alleles were detected in MNU-treated K, KP and KPL cell lines. The VAFs of the Kras mutation were around 0.4–0.6 in all cell lines, indicating that each cell line maintained the heterozygous Kras mutation. We then assessed the evolution of mutations within each family of cell lines with increased exposure to MNU, and detected mutations that were shared among parental, 3M, 5M and 7M cells within each genetic background, as well as private mutations which were specific to each cell line (Fig. 1d). The presence of both private and shared mutations in each family of cell lines provides a unique opportunity to evaluate the immune editing of these mutations in response to therapy in syngeneic studies.
Higher TMB results in decreased tumor growth
To determine the effect of increased TMB on in vivo tumor growth, we screened each family of cell lines with varying TMB in syngeneic models. Across all genomic backgrounds, we observed diminished in vivo tumor growth with incremental increases in TMB (Fig. 2a). Increasing the number of injected cells led to robust tumor growth of K-3M, KP-3M, KPL-3M and KPL-5M cells, while other lines, namely K-5M, K-7M, KP-5M, KP-7M, KPL-7M, were rejected or displayed diminished growth rates (data not shown). Evaluation of in vitro growth rates of cell lines with varying TMB within each family revealed minimal differences (Supplemental Figure S3). These results indicate that diminished in vivo growth associated with increased TMB is likely immune-mediated. To further confirm this finding, we evaluated the in vivo growth of the K, KP and KPL parental cells, and their associated 7M counterparts in immunocompromised SCID mice which lack T and B cells (Fig. 2b). We observed similar tumor growth rates of K-7M and KP-7M compared to their parental counterparts, while KPL-7M showed minor but statistically significant reduction in tumor growth compared to parental KPL cells. Given that there was no difference in growth rates of KPL-P and KPL-7M in vitro (Supplemental Figure S3), the slightly reduced growth rate of KPL-7M in vivo could possibly be related to other immune-mediated pathways such as natural killer cells. These results suggest that the decreased tumor growth rates associated with high TMB in immunocompetent mice are predominantly due to host adaptive immune responses.
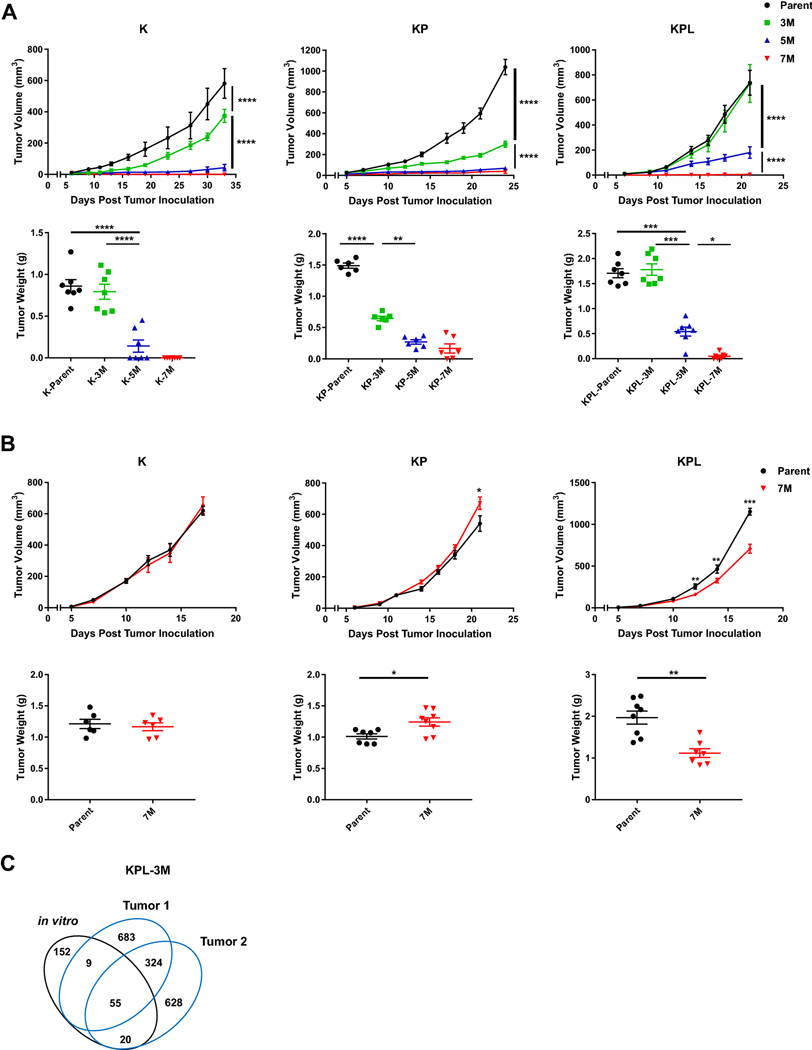
Fig. 2.
In vivo tumor growth in immunocompetent and SCID mice. a Within each family of cells, the Parent, 3M, 5M, and 7M cells were inoculated SC in immunocompetent mice [K (2 × 106) cells in 129-E mice; KP (8 × 105) cells in FVB mice; KPL (1 × 105) cells in FVB mice] and tumor growth was measured with caliper. Growth curves and corresponding tumor weights after euthanasia are presented. b Same as in a except Parent and 7M cells were inoculated SC in SCID mice [K (2 × 106) cells; KP (8 × 105) cells; and KPL (1 × 105) cells]. Data are representatives of at least two biological replicates of 6–10 mice per group. c Venn diagram of shared and private mutations of the KPL-3M cell line and two subcutaneous KPL-3M tumors from immunocompetent mice at day 25 post inoculation. P values were determined by two-tailed non-paired Student’s t test for pairwise comparison and two-way ANOVA with Tukey post-test for time-associated comparison among multiple groups. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001
Next, we assessed the stability of the TMB in our syngeneic model system to ensure that in vivo host immune editing does not result in outgrowth of tumors with low TMB. We utilized KPL-3M as a representative model and performed WES of KPL-3M subcutaneous tumors in immunocompetent mice at day 25. We observed that subcutaneous KPL-3M tumors maintained 30% of the mutations present in KPL-3M cells prior to in vivo mouse inoculation (Fig. 2c). These represent stable mutations that are not edited by host immune cells. We also observed emergence of new mutations which likely result from the expansion of minor subclones that are below the detection threshold in KPL-3M cells (Fig. 2c). Overall, approximately 30% of total mutations were shared between two independent tumors (Fig. 2c). Taken together, these data suggest that the KPL-3M tumors maintain high TMB in vivo with a considerable overlap in their mutational profiles.
High TMB is associated with increased local and systemic tumor-specific T cells
To define the immune responses induced by high TMB in each genetic background, we utilized cells with robust in vivo growth and moderate mutation burden, namely K-Parent, K-3M, KP-Parent, KP-3M, KPL-Parent, KPL-3M, and KPL-5M. We first evaluated the lymphoid compartment of the TME. We observed a significant increase in the number of tumor-infiltrating lymphocytes (TILs) and an increase in CD8+ to regulatory T (Treg) cell ratio in the TME with increased TMB in each genetic background (Fig. 3a). In addition, a higher percentage of infiltrating CD8+ T cells in the tumors with high TMB expressed the proliferation marker Ki-67 compared to their respective parental tumors.
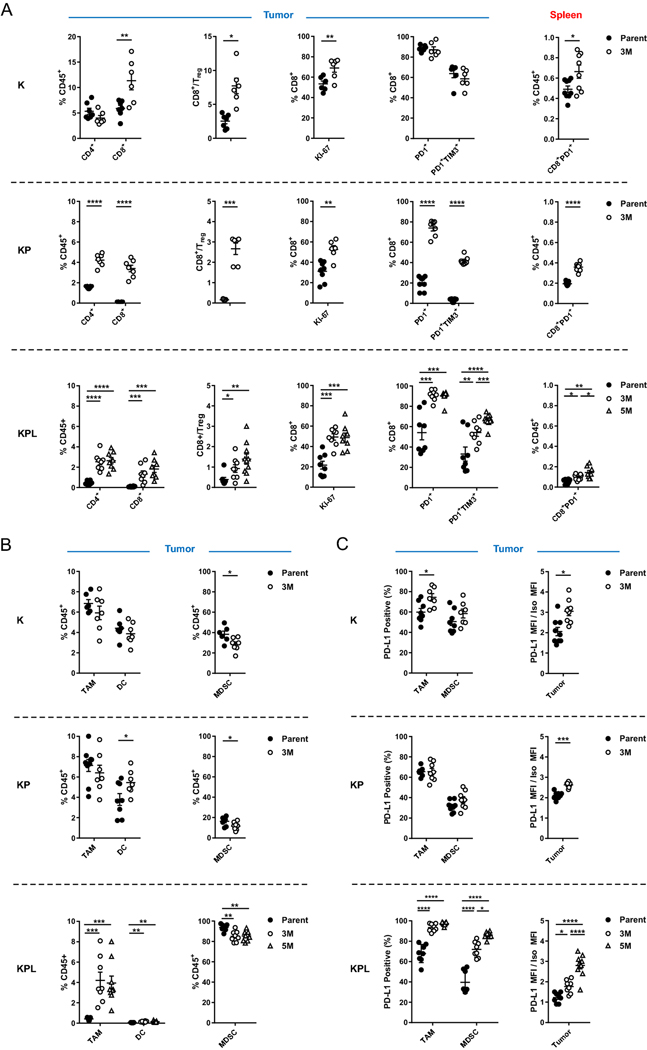
Fig. 3.
Distinct immune phenotypes of murine models revealed by FACS. On day 14–16 post-tumor inoculation [2 × 106 K-Parent and K-3M cells in 129-E mice; 8 × 105 KP-Parent and 2 × 106 KP-3M cells in FVB mice; 1 × 105 KPL-Parent, 1.5 × 105 KPL-3M, and 3 × 105 KPL-5M cells in FVB mice], tumors and spleens were harvested and analyzed by FACS. a Lymphoid compartment. b Myeloid compartment. c PD-L1 expression. Data are representatives of at least two biological replicates of 6–10 mice per group. P values were determined by P values were determined by two-tailed non-paired Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001
Next, we evaluated the expression of the early activation/exhaustion marker PD-1 on TILs and observed higher expression of PD-1 on CD8+ T cells in KP and KPL tumors with high TMB compared to their parental counterparts (Fig. 3a). Studies reveal that tumor-specific CD8+ TILs in human cancers express high levels of PD-1 and that this phenotype can identify the diverse repertoire of clonally expanded tumor-reactive T cells. Thus, our results suggest that increased TMB in the KP and KPL models results in increased tumor-specific CD8+ T cell responses [16, 17]. In parallel, we detected increased co-expression of the checkpoint TIM-3, a marker of increased T cell exhaustion with prolonged antigen exposure, on PD-1+CD8+ T cells in KP and KPL tumors with high TMB compared to their parental counterparts. However, we observed no difference in tumor CD8 + T cell exhaustion between K-parent and K-3M tumors. This observation is likely due to increased immunogenicity of the K-parent tumors, which contain high TILs and increased CD8 + T cell exhaustion at baseline and exhibit slow in vivo growth with modest sensitivity to anti-PD-1 therapy. These results support the hypothesis that a higher TMB results in increased tumor-reactive PD-1+ T cells within the TME, which become exhausted with persistent antigen stimulation.
Peripheral tumor neoantigen-specific T cells which overlap with clonal tumor-specific TILs have also been identified in circulating PD-1+CD8+ T cells in melanoma patients [18]. Therefore, we evaluated the lymphoid compartment within the spleen of tumor-bearing mice, including expression of PD-1 on CD8 + T cells (Fig. 3a). We observed an increase in splenic PD-1+CD8+ T cells in mice bearing K, KP, and KPL tumors with high TMB compared to the parental counterparts, including a statistically significant increase in mice bearing KPL-5M tumors compared to those bearing KPL-3M tumors (Fig. 3a). These data suggest that increased TMB results in increased systemic tumor-specific T cells in our murine models.
Next, we evaluated the myeloid compartment of the TME and observed TMB-mediated changes in the immune phenotypes shared across all genetic backgrounds, as well as marked differences specific to KPL cells (Fig. 3b and Supplemental Figure S4A). High TMB was associated with a significant increase in the professional antigen-presenting dendritic cells (DCs) in the KP and KPL tumors, with no differences observed in K tumors. We observed no differences in the number of tumor-associated macrophages (TAMs) in K-3M and KP-3M compared to the respective parental tumors, but we observed an increase in TAMs in KPL-3M and KPL-5M tumors compared to KPL-Parent. We evaluated changes in the MDSCs and found that high TMB was associated with decreased MDSCs across all genetic backgrounds. KPL tumors contained a significantly higher percentage of MDSCs (over 80% of CD45+ cells), which were predominantly G-MDSCs expressing the neutrophil marker Ly6G (Fig. 3b and Supplemental Figure S4B). We observed a similar phenotype in the spleen where KPL tumor-bearing mice had a significantly higher percentage of G-MDSCs compared to mice bearing K and KP tumors (Supplemental Figure S4C). This immune phenotype in KPL models is consistent with studies in Kras-mutant murine and KRAS-mutant human NSCLC where LKB1 loss is associated with a T cell-suppressed and neutrophil-enriched TME [19–21].
We further evaluated the expression of PD-L1 by tumors and the myeloid cells in the TME (Fig. 3c). We observed an increase in PD-L1 expression on TAMs in K and KPL tumors with increased TMB compared to their parental counterparts but no difference between KP-3M and KP-Parent. We observed increased PD-L1 expression on MDSCs in the KPL tumors with higher TMB, with the greatest expression observed in KPL-5M, but no difference was detected within the K and KP genetic background. Increased PD-L1 expression was also observed on tumors with an increased TMB in each genetic background. Taken together, the observed overall trend of increased PD-L1 expression associated with increasing TMB implies amplified adaptive immune resistance within the TME.
Anti-PD-1 responses in cell lines with high TMB recapitulate the therapeutic vulnerabilities of KRAS-mutant human NSCLC
We evaluated the efficacy of PD-1 blockade in K-3M, KP-3M and KPL-3M tumors with increased TMB (Fig. 4a and Supplemental Figure S5A). Anti-PD-1 therapy resulted in robust anti-tumor responses with an eradication of 33% of K-3M tumors. Similarly, 44% of KP-3M tumors were rejected and others stabilized in response to anti-PD-1. In contrast, anti-PD-1 efficacy was limited in KPL-3M tumors where PD-1 blockade resulted in reduced tumor growth without a complete rejection. This result is in agreement with the recent findings in human KRAS-mutant lung adenocarcinoma in which LKB1 loss was shown to be a major driver of primary resistance to PD-1 blockade [20]. We next assessed the efficacy of PD-1 blockade in mice bearing KPL-5M tumors and observed significant anti-tumor responses with the rejection of approximately 50% of tumors (Fig. 4b). These data suggest that the increased TMB of KPL-5M tumors could overcome the immunosuppressed TME and enhance responses to PD-1 blockade. This is in agreement with our immunophenotyping results of the KPL family of tumors in which mice bearing KPL-5M tumors possessed the highest number of local and systemic activated PD1+ CD8+ T cells (Fig. 3a). PD1+ CD8+ T cells have been shown to contain pools of tumor neoantigen-specific T cells that can be reinvigorated following PD-1 blockade [22, 23]. The mice bearing KPL-5M tumors, that had a complete anti-tumor response to anti-PD1, were subsequently rechallenged with KPL-5M cells. In response to the rechallenge, we observed an initial tumor growth followed by spontaneous rejection of all tumors (Fig. 4c), indicating the establishment of systemic anti-tumor immunity in response to PD-1 blockade in mice bearing KPL-5M tumors.
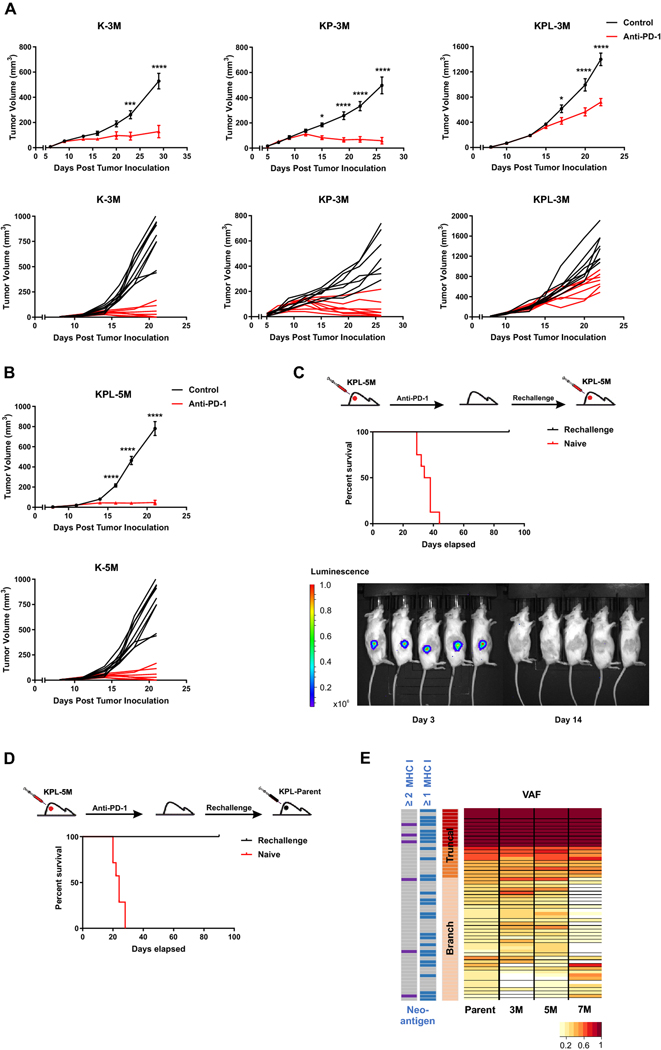
Fig. 4.
High TMB results in increased efficacy of anti-PD-1 therapy. a After SC tumor inoculation [K-3M (2 × 106) cells in 129-E mice; KP-3M (2 × 106) cells in FVB mice; KPL-3M (1.5 × 105) cells in FVB mice], mice bearing < 50mm3 tumors (∼ day 7) were treated with (i) isotype control, (ii) anti-PD-1 (200 μg/dose 3 times weekly for 4 doses), and tumor growth was measured with caliper. Results are representatives of at least two biological replicates of 6–10 mice per group. b Same experimental design as a except that KPL-5M (3 × 105) cells were utilized for SC tumor inoculation. c FVB naïve mice and mice that previously eradicated KPL-5M tumors in response to PD-1 blockade were inoculated SC with KPL-5M (3 × 105) and tumor growth was measured with bioluminescence imaging on day 3 and day 14. Survival curve is presented. Data is representatives of two biological replicates of 5–6 mice per group. d FVB naïve mice and FVB mice that previously eradicated KPL-5M tumors in response to PD-1 blockade were inoculated SC with KPL-Parent (2 × 105). Survival curve is presented. Data is representatives of two biological replicates of 5–6 mice per group. e Representation of the frequency of the mutations in the KPL-Parent tumors shared by KPL-3M, KPL-5M, or KPL-7M. The predicted neoantigens are presented based on MHC-I binding avidity by at least one or two MHC-I alleles (two left columns). P values were determined by two-way ANOVA with Tukey post-test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001
Given that KPL-5M shares 47 truncal and branch mutations with the KPL-Parent (Fig. 1d), we assessed whether the mice that had eradicated the KPL-5M tumors following anti-PD-1 treatment could reject the parental tumors by inoculating the mice with KPL-Parent cells 3 months after the initial rejection (Fig. 4d). All of the mice eliminated the KPL-Parent tumors after an initial growth, while the naïve control mice succumbed to implantation of KPL-Parent tumors in less than 30 days. Computational analysis of putative neoantigens in the KPL-Parent revealed 11 truncal and 17 branch neoantigens which were shared with KPL-3M, KPL-5M or KPL-7M (Fig. 4e and Supplemental Figure S5B). These results indicate the presence of tumor-specific memory T cells against shared neoantigen(s) between KPL-5M and KPL-Parent tumors in anti-PD-1 treated mice that had eradicated KPL-5M tumors. Despite this finding, T cell responses against these shared neoantigen(s) are not sufficient to eradicate KPL-Parent tumors in naïve mice treated with PD-1 blockade (Fig. 1a). This may be due to profound immunosuppression in the TME of KPL-Parent tumors that represses the initial host anti-tumor T cell responses (Fig. 3). In contrast, eradication of KPL-P tumors in the rechallenge experiments may be predominantly mediated by memory T cells, which can generate a rapid recall response to secondary challenge that overcomes the immunosuppressive TME. The presence of shared neoantigen(s) in these isogenic cell lines with varying TMB provides a unique opportunity to investigate immune responses against truncal and branch mutations in the context of TMB-associated changes in the TME.
In summary, we report novel Kras-mutant murine models of NSCLC bearing common driver gene alterations and increased TMB (Supplemental Table S2). Although the nature of the additional somatic mutations induced by MNU may not fully recapitulate the spectrum of tobacco-related mutations observed in human KRAS-mutant NSCLC, these murine models are clinically relevant because they possess the dominant driver mutations of KRAS-mutant NSCLC that determine distinct TMEs and clinical phenotypes as well as passenger mutations that can elicit adaptive immune responses. In contrast to existing Kras-mutant GEMMs of NSCLC that possess few mutations and have limited utility in immunotherapy studies, our models with increased TMB recapitulate the therapeutic vulnerabilities to anti-PD-1 which mirrors that of human KRAS-mutant NSCLC. The KPL-3M model with co-occurring Kras and Lkb1 mutations, neutrophil-enriched TME, physiologically relevant TMB, and limited efficacy to anti-PD-1 serves as a clinically relevant model for preclinical immunotherapy studies, given loss of LKB1 is a dominant driver of resistance in human NSCLC. We anticipate that these novel immunogenic murine models will facilitate the development of future immunotherapies for NSCLC.
Supplementary Material
Acknowledgements
The authors thank Lauren Winter and Tamara Silva for the administrative support.
Funding This research was supported in part by funding from the National Cancer Institute (1U01-CA196408), National Heart Lung and Blood Institute (T32-HL072752), Medical Research Funds from the Department of Veteran Affairs, and NIH/NCATS UL1TR001881. The UCLA Jonsson Comprehensive Cancer Center (JCCC) and Center for AIDS Research Flow Cytometry Core is supported by the National Institutes of Health (NIH; #CA-16042 and #AI-28697).
Footnotes
Availability of data and materials The authors confirm that the data supporting the findings of this study are available within the article and its supplementary material.
Code availability Not applicable.
Compliance with ethical standards
Ethics approval Research involving animals were reviewed and approved by the UCLA Animal Research Committee prior to starting the project. The committee ensures that the study is ethical and that the animals are treated humanely.
Consent to participate This manuscript does not involve human participants, human data or human tissue.
Consent for publication This manuscript does not involve human participants, human data or human tissue.
Conflict of interest Steven M. Dubinett is an advisory board member for EarlyDx Inc., T-Cure Bioscience Inc., LungLife AI Inc., and Johnson and Johnson Lung Cancer Initiative.
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/s00262-020-02837-9.
References
- 1.Gandhi L, Rodríguez-Abreu D, Gadgeel S et al. (2018) Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med 378:2078–2092. 10.1056/NEJMoa1801005 [DOI] [PubMed] [Google Scholar]
- 2.Rizvi NA, Hellmann MD, Snyder A et al. (2015) Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348:124–128. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tumeh PC, Harview CL, Yearley JH et al. (2014) PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515:568–571. 10.1038/nature13954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garon EB, Rizvi NA, Hui R et al. (2015) Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 372:2018–2028. 10.1056/NEJMoa1501824 [DOI] [PubMed] [Google Scholar]
- 5.Gettinger SN, Choi J, Mani N et al. (2018) A dormant TIL phenotype defines non-small cell lung carcinomas sensitive to immune checkpoint blockers. Nat Commun 9:3196. 10.1038/s41467-018-05032-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGranahan N, Furness AJ, Rosenthal R et al. (2016) Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351:1463–1469. 10.1126/science.aaf1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawrence MS, Stojanov P, Polak P et al. (2013) Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499:214–218. 10.1038/nature12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skoulidis F, Byers LA, Diao L et al. (2015) Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov 5:860–877. 10.1158/2159-8290.CD-14-1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westcott PM, Halliwill KD, To MD et al. (2015) The mutational landscapes of genetic and chemical models of Kras-driven lung cancer. Nature 517:489–492. 10.1038/nature13898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung W-J, Daemen A, Cheng JH et al. (2017) Kras mutant genetically engineered mouse models of human cancers are genomically heterogeneous. Proc Natl Acad Sci 114:E10947–E10955. 10.1073/pnas.1708391114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McFadden DG, Politi K, Bhutkar A et al. (2016) Mutational landscape of EGFR-, MYC-, and Kras-driven genetically engineered mouse models of lung adenocarcinoma. Proc Natl Acad Sci USA 113:E6409–E6417. 10.1073/pnas.1613601113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olson B, Li Y, Lin Y, Liu ET, Patnaik A (2018) Mouse models for cancer immunotherapy research. Cancer Discov 8:1358–1365. 10.1158/2159-8290.CD-18-0044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wislez M, Spencer ML, Izzo JG et al. (2005) Inhibition of mammalian target of rapamycin reverses alveolar epithelial neoplasia induced by oncogenic K-ras. Cancer Res 65:3226–3235. 10.1158/0008-5472.CAN-04-4420 [DOI] [PubMed] [Google Scholar]
- 14.DeVette CI, Andreatta M, Bardet W, Cate SJ, Jurtz VI, Jackson KW, Welm AL, Nielsen M, Hildebrand WH (2018) NetH-2pan: a computational tool to guide MHC peptide prediction on murine tumors. Cancer Immunol Res 6:636–644. 10.1158/2326-6066.CIR-17-0298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A (2017) Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168:707–723. 10.1016/j.cell.2017.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmadzadeh M, Johnson LA, Heemskerk B, Wunderlich JR, Dudley ME, White DE, Rosenberg SA (2009) Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood 114:1537–1544. 10.1182/blood-2008-12-195792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gros A, Robbins PF, Yao X et al. (2014) PD-1 identifies the patient-specific CD8(+) tumor-reactive repertoire infiltrating human tumors. J Clin Invest 124:2246–2259. 10.1172/JCI73639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gros A, Parkhurst MR, Tran E et al. (2016) Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med 22:433–438. 10.1038/nm.4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koyama S, Akbay EA, Li YY et al. (2016) STK11/LKB1 deficiency promotes neutrophil recruitment and proinflammatory cytokine production to suppress T-cell activity in the lung tumor microenvironment. Cancer Res 76:999–1008. 10.1158/0008-5472.CAN-15-1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skoulidis F, Goldberg ME, Greenawalt DM et al. (2018) STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov 8:822–835. 10.1158/2159-8290.CD-18-0099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biton J, Mansuet-Lupo A, Pecuchet N et al. (2018) TP53, STK11, and EGFR mutations predict tumor immune profile and the response to anti-PD-1 in lung adenocarcinoma. Clin Cancer Res 24:5710–5723. 10.1158/1078-0432.CCR-18-0163 [DOI] [PubMed] [Google Scholar]
- 22.Yost KE, Satpathy AT, Wells DK et al. (2019) Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat Med 25:1251–1259. 10.1038/s41591-019-0522-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamphorst AO, Pillai RN, Yang S et al. (2017) Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc Natl Acad Sci USA 114:4993–4998. 10.1073/pnas.1705327114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.