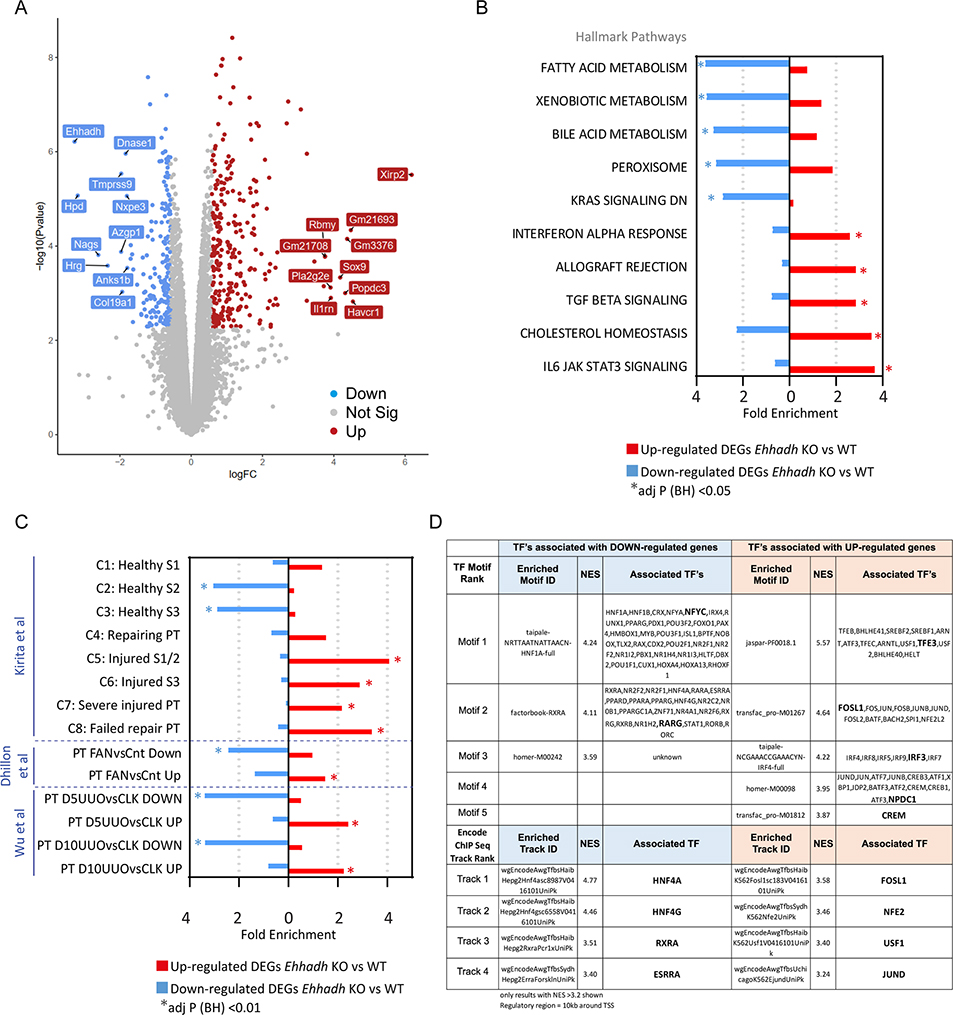
Figure 2. Transcriptomics of male Ehhadh KO kidneys.
A) Volcano plot with significantly differentially expressed genes (DEGs) highlighted. Decreased genes (down) are highlighted in blue, increased genes (up) are highlighted in red. Gene names indicate the top 10 significant up and down genes by logFC using an adj p < 0.05. Genes that were not statistically significant are represented in grey. B) Top 10 pathways (by fold enrichment) after pathway enrichment analysis of genes either significantly up- or down- regulated in Ehhadh KO mice versus WT according to the Hallmark database. Values represent the fold enrichment and significance is indicated as * (adj p<0.05). Full table of results are in Table S1B and S1C. C) Results of enrichment analysis of genes either up- or down- regulated in Ehhadh KO mice versus WT, in gene sets curated from three independent murine proximal tubule (PT) acute kidney injury models (see Methods). Values represent the fold enrichment and significance is indicated as * (adj p<0.01). Full table of results are in Table S1D. D) Predicted transcriptional regulators as identified by Iregulon through Motif or Encode ChIP-seq enrichment analysis for the genes either up- or down- regulated in Ehhadh KO mice versus WT kidney samples. Only results with normalized enrichment score (NES) >3.2 are shown. Full table of results are in Table S1E. The bolded transcription factor (TF) represents the most likely TF associated with the enriched motif. Human orthologs of the murine DEGs (at adj p<0.05) were the input for B), C) and D).