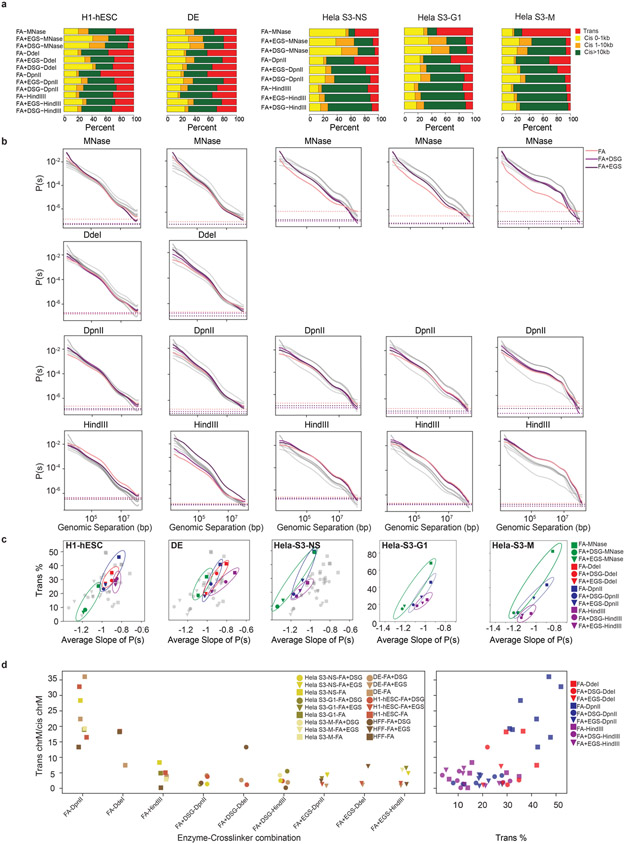
Extended Data Fig. 2. Cis and trans contact frequency differ between protocols.
a. The number of valid pairs in each of the 12 protocols applied to H1-hESC, DE, HeLa-S3-NS, HeLa-S3-G1 and HeLa-S3-M cells partitioned by genomic distances.
b. Distance dependent contact probability of 12 protocols ordered as in (a), partitioned by fragmenting nucleases used (gray lines indicate all datasets, colored lines indicate datasets generated with the nucleases indicated for each plot).
c. The relationship between the trans percent and the average slope of the distance dependent contact probability for the 12 protocols ordered as in Extended Data Fig. 2a.
d. Quantification of protocol introduced noise as defined by inter-mitochondrial interactions (chrM with chr1-22), normalized by intra-mitochondrial (chrM with chrM) interactions.