Abstract
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), like other coronaviruses, relies on a flexible array of entry mechanisms, driven by the spike (S) protein. Entry is dependent on proteolytic priming, activation, and receptor binding; all of which can be variable, dependent on context. Here we review the implications of the complexity of SARS-CoV-2 entry pathways on entry assays that then drive our understanding of humoral immunity, therapeutic efficacy, and tissue restriction. We focus especially on the proteolytic activation of SARS-CoV-2 spike and how this constellation of proteases lends deeper insight to our understanding of arising variants and their putative role transmission or variable pathogenicity in vivo. In this review, we argue for better universal standards to assay virus entry as well as suggest best practices for reporting viral passage number, the cell line used, and proteases present, among other important considerations.
Keywords: SARS-CoV-2, proteases, Furin, TMPRSS2, virus entry, variants
Introduction
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) emerged in humans in late 2019, being first identified as the causative agent of coronavirus disease 2019 (COVID-19) in early 2020 [1,2]. As the COVID-19 pandemic raged, scientists responded with an unprecedented surge of biomedical research activity to better understand the pathobiology of SARS-CoV-2, identify putative therapeutics, and develop vaccines [3]. Many of these efforts rely on understanding the mechanics of virus entry. Our search for a correlate of immunity also relies on how we quantify neutralizing antibodies that block virus entry. Many SARS-CoV-2 variants of concern/interest (VOC/VOI) have mutations that affect the efficiency of virus entry or evade antibodies that would otherwise block virus entry. Here, we bring to the forefront considerations for studying SARS-CoV-2 entry, specifically focusing on proteases, cleavage variants, and their implications for the field and the assessment of putative therapeutics.
Considerations for SARS-CoV-2 Entry
Like other coronaviruses, SARS-CoV-2 entry is facilitated by the surface glycoprotein S, a class I fusion protein. The protein is comprised of an N-terminal S1 and a C-terminal S2 subunit. Notably, the S1 domain contains the receptor binding domain (RBD), which is responsible for recognition and binding to the angiotensin-converting enzyme 2 (ACE2) receptor on host cells [4–6]. The S2 domain contains the S2’ cleavage site, the fusion peptide (FP) directly adjacent to S2’, the two heptad repeats (HR1 and HR2), the transmembrane domain (TM), and the cytoplasmic tail (CT) [7] (Fig. 1A). S2 is primarily responsible for membrane fusion between the virus and the host cell—a process that requires proteolytic activation followed by significant receptor induced conformational changes [8–10]. SARS-CoV-2 S also contains an insertion at S1/S2 that results in a multibasic cleavage site, 681-PRRAR^S-686, making it unique among sarbecoviruses in betacoronavirus lineage B (Fig. 1A/B).
Figure 1. SARS-CoV-2 entry is dependent on proteolytic cleavage of the spike glycoprotein mediated through early or late entry pathways.
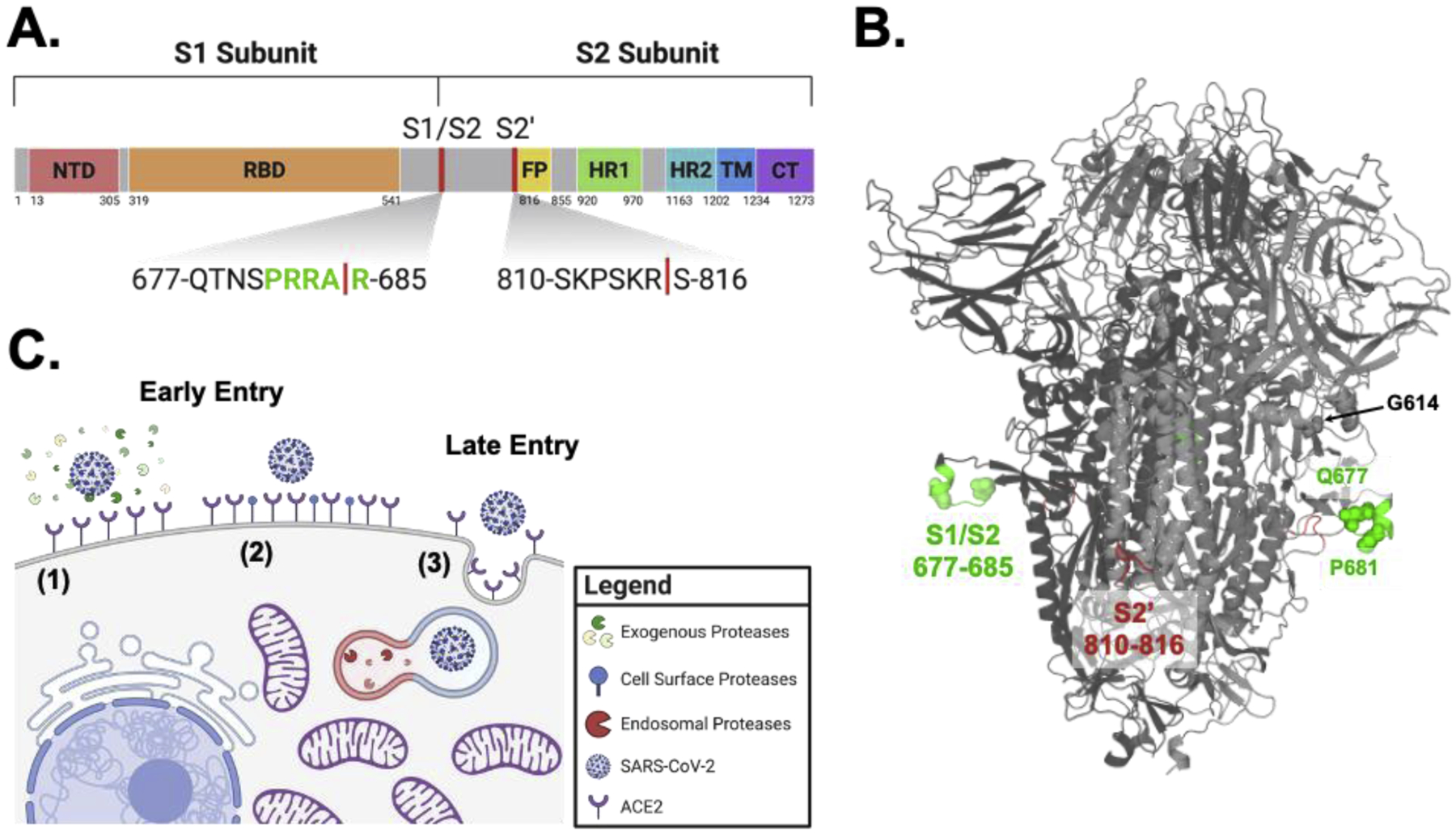
(A) Schematic diagram of spike glycoprotein highlighting S1/S2 and S2’ cleavage sites. Uniport SARS-CoV-2 spike annotations were used and schematic was generated in Biorender. (B) Structure of spike glycoprotein depicting cleavage locations and select mutants. SARS-CoV-2 structure generated using SWISS-MODEL to depict the S1/S2 cleavage sites and annotated using Pymol. S1/S2 region is depicted in green and S2’ region is in red. Spheres depict the location of select mutations discussed in the text: G614, Q677, and P681. (C) Proteolytic activation of SARS-CoV-2 spike mediates early and late entry pathways. Pathways 1 and 2 show early entry at the cell surface facilitated by exogenous or cell surface proteases, respectively. Pathway 3 shows late entry facilitated by endosomal proteases such as cathepsin L. Model generated using Biorender and adapted from Oguntuyo and Stevens et al.
SARS-CoV-2 S is proteolytically activated in two sequential steps: 1) cleavage at the S1/S2 site, then 2) subsequent cleavage at S2’ (Fig. 1C) [11,12]. The purpose of the first of these two cleavage events is to separate the component primarily responsible for receptor binding (S1) and the component primarily containing the fusion machinery (S2), resulting in a non-covalent association between S1 and the transmembrane-anchored S2. This first cleavage event at S1/S2 is distant from the fusion peptide and distant from any significant region of hydrophobic peptides—a characteristic making spike distinct from some other class I fusion proteins such as hemagglutinin from influenza or gp160 from HIV where the cleavage sites are directly adjacent to the fusion peptides. While S1/S2 cleavage does not appear to reveal a fusion peptide motif, it does play the role of priming spike for S2’ cleavage. Cleavage at the S1/S2 site often occurs during spike processing within producer cells and improves S2’ site accessibility and cleavage efficiency [13]. The second cleavage event at S2’ is primarily then to reveal the fusion peptide. The timing of this cleavage is likely regulated to prevent premature fusion protein triggering, enabling more efficient productive membrane fusion.
The presence of these two distinct cleavage sites and the potential for two distinct cleavage events to occur, facilitated by different proteases, means that faithful representation of the biology of SARS-CoV-2 entry requires many moving pieces. S1/S2 cleavage is thought to occur via furin-like proteases, cathepsins, and other host proteases [8,9,13–18] while S2’ cleavage can happen either in the extracellular space or at the cell surface by a wide variety of serine proteases such as trypsin, neutrophil elastase (NE), and TMPRSS2 [19–21]. There also exist a number of additional potential cleavage sites, some of which may play a role in viral entry or even result in spike inactivation [18]. The system that regulates SARS-CoV-2 entry is further complicated by physiologically relevant protease inhibitors such as alpha-1 antitrypsin [22] that antagonizes many of the abovementioned proteases at their site of action. We also see that in the absence of S1/S2 cleavage, S2’ activation is still possible by cathepsins like cathepsin L via the endocytic pathway [17]. In summary, SARS-CoV-2 can enter and fuse at the plasma membrane or in the endosomes, which are commonly referred to as the early or late pathways, respectively. The former requires S1/S2 cleavage then S2’ cleavage by extracellular or cell surface serine proteases like trypsin, NE, or TMPRSS2, while the latter is driven by endosomal proteases like cathepsin L (Fig. 1C) [19,23–25].
The tissue specific nature of proteases, inhibitors, and cellular receptors may also play a role in the variable selective pressures faced by the virus, even within an individual host. SARS-CoV-2 sequence mutations have been shown to occur in vivo with differential sequence selection in different tissue types of human post mortem samples. This raises the possibility that tissue restriction or tissue-specific viral growth kinetics may be subject to change over the course of infection as selective pressure is exerted on the virus [26].
The interplay between tissue-type and proteolytic activation has been shown to be important in determining cell and tissue-specific viral tropism. For example, in the oral cavity, salivary proteases may help provide favorable conditions for entry and replication [27]. There, SARS-CoV-2 is activated by a number of tissue-specific proteases such as TMPRSS2, TMPRSS4 and TMPRSS11D; as well as endosomal proteases like CTSB, CTSL, BSG, and furin [28]. It is therefore fortunate that, given the high concentration of a wide range of activating proteases, both vaccinated and convalescent patients display high levels of antibodies in the saliva [29]. Single cell RNA expression experiments have shown that the oral cavity is but one example of the ways in which a variety of host entry factors [28], including tissue-specific proteases, help modulate cell tropism [30].
Through all this, we see that SARS-CoV-2 entry is driven, in part, by cell or tissue type specific proteases, as well as proteases that can accumulate in the cognate extracellular milieu [30–32]. Here, we discuss the interplay between proteolytic activation and genomic sequence variation. We argue that the proper interpretation of SARS-CoV-2 entry assays in various contexts need to take into account this complex interplay.
How SARS-CoV-2 Entry is Assessed
Entry assays are used to determine how well particular spike proteins mediate entry, as well as to assess the breadth and potency of neutralizing antibody responses in convalescent or vaccine sera. The latter is particularly important as we seek a correlate of immunity that reflects the efficacy of a given vaccine.
Differential proteolytic activation of SARS-CoV-2 spike can lead to divergent entry pathways (Fig. 1C) that impinge on the infectivity of a given virus stock and its sensitivity to neutralizing antibodies. Understanding the parameters that govern SARS-CoV-2 entry is of vital importance in interpreting the results of different virus entry and neutralizing assays. Table 1 shows a representative selection of studies utilizing entry assays to look at vaccine efficacy, screen for convalescent plasma, and investigate the impact of spike variants on the neutralizing activity of post-vaccine sera. In each category, we chose the earliest major study that was eventually published in a peer-reviewed journal, and included international studies whenever possible. All of the studies describing vaccine efficacy use at least one form of live virus neutralization assay. A few were complemented by a pseudotyped virus neutralization assay (Fig. 2 and Table 1). Notably, none of these studies utilized cell lines containing TMPRSS2 nor did they supplement with trypsin or any other additional proteases. Of the studies on convalescent plasma therapy, only one utilized a cell line expressing TMPRSS2 (293T-ACE2-TMPRSS2) in the context of pseudotyped viruses [33] while one other used trypsin in their microneutralization assays (Table 1, #11 and #13, respectively) [34]. Strikingly, all the live virus neutralization assays indicated in Table 1 were performed in Vero cells, where entry occurs exclusively via the late pathway and is dependent on endosomal proteases such as cathepsin L.
Table 1. Assay characteristics of select vaccine, convalescent plasma, and variant entry inhibition studies.
To facilitate interpretation of the studies included in these tables, the characteristics of the studies have been color coded within each category and further annotated for additional information.
| Study type/Reference | Assay type | Cell line | Media | Serum and Heat Inactivation (HI) | Exogenous protease | reference | |
|---|---|---|---|---|---|---|---|
| Vaccine | moderna | pseudotyped lentivirus and wildtype SARS-CoV-2 PRNT | 293T-ACE2 of lenti virus; vero E6 for plaques | DMEM overlay | sera; HI 56°C for 45mins | no | NEJM Epub Jul2020 |
| novavax | live virus microneut | vero E6 | DMEM+10%FBS | sera; HI 56°C for 30mins | no | NEJM Epub Sept 2020 | |
| pfizer | live virus NG | grown in veroE6, infected vero CCL81 | DMEM+2% FBS | sera; HI 56°C for 30mins | no | Nature | |
| sputnik | live virus CPE | vero E6 | DMEM+2%FBS | sera; HI 56°C for 30mins | no | Lancet | |
| astrazeneca | live virus plaques/PRNT and lentivirus | vero E6 or 293Ts transfected w/ACE2 | MEM+0.5%FCS and others not reported | sera; HI 56°C for 30mins-1hr | no | Lancet | |
| johnson and johnson | live virus plaques | vero E6 | not reported | sera; HI, condition not described | no | NEJM | |
| sinovac | VSVΔG pseudovirus and wt virus micro CPE | huh7 cells for VSVΔG?; vero E6 for live virus | DMEM + 2% new born calf serum | sera; HI, 56°C for 30mins | no | Lancet ID | |
| bharat biotech | live virus PRNT and microneut | vero CCL81 for PRNT | not reported | sera; HI, 56°C for 30mins | no | Lancet ID | |
| Convalescent plasma | Convalsecent plasma | Competitive ELISA | N/A | N/A | serum; not reported | no | NEJM |
| Convalsecent plasma | VSVΔG pseudotyped particles | 293T-ACE2+TMPRSS2 | DMEM+10%FBS | plasma; not reported | no | NEJM | |
| Convalsecent plasma | live virus microneut | vero E6 cells | MEM+2% FBS | serum; HI 56°C for 1hr | no | Nature | |
| Convalsecent plasma | live virus microneut | vero E6 cells | Eagle’s MEM | serum; not reported | trypsin | Haematologica | |
| Convalsecent plasma | live virus plaques | vero E6 cells | not reported | serum; HI 56°C for 30mins | no | PNAS | |
| Convalsecent plasma | live virus CPE | vero cells | MEM | serum; HI 56°C for 30mins | no | JAMA | |
| Convalsecent plasma | live virus CPE | vero CCL81 | not reported | serum; HI 56°C for 30mins | no | JAMA | |
| Convalsecent plasma | live virus PRNT | vero E6 cells | DMEM+1%FBS | serum; HI 56°C for 30mins | no | medRxiv | |
| Convalsecent plasma | replication competent VSV | vero cells | MEM+10%FBS | serum; HI 56°C for 30mins | no | JCI-Insight | |
| variants | B.1.1.7 and B.1.351 | live virus microneutralization, VSVΔG | vero E6 cells | EMEM 7.5% FCS | serum; HI, condition not described | no | medRxiv |
| B.1.1.7 and B.1.351 and others | lentivirus | 293T-ACE2 | not reported | serum; not reported | no | Cell | |
| B.1.351 | lentivirus and live virus focus forming assay | 293T-ACE2 or vero E6 cells | DMEM+10% FBS | serum; HI 56°C for 30mins and clarified 5mins at 10K rcf | no | NEJM | |
| B.1.1.7 and B.1.351 and P.1 | VSVΔG pseudotyped particles | vero E6 cells | DMEM+10%FBS | serum; HI 56°C for 30mins | no | Cell | |
| B.1.1.7 | live virus microneutralization | vero E6 cells | MEM+0.5%FCS | serum; HI 56°C for 30mins | no | Lancet Pre-Print | |
| B.1.1.7 and B.1.351 and E484K | replication competent | 293T-ACE2-TMPRSS2 | DMEM+10%FBS | serum; HI 56°C for 30mins | no | medRxiv |
Abbreviations for live or pseudotyped virus: P (Plaque Reduction Neutralizing Titers or PRNT), F (focus forming units or FFUs), RG (Reporter Genome), MN (Microneutralization), LV (Lentivirus), ΔG (VSVΔG), rV (replication competent VSV), or other. Cell line abbreviations: Vero-C/E (CCL81 or E6; when absent, this indicates that the study did not denote the clone utilized), 293T-A (ACE2 stably expressing; A# denotes 293Ts transfected with ACE2 prior to infection), 293T-AT (ACE2 and TMPRSS2).
Figure 2. Tools to assess SARS-CoV-2 entry inhibition.
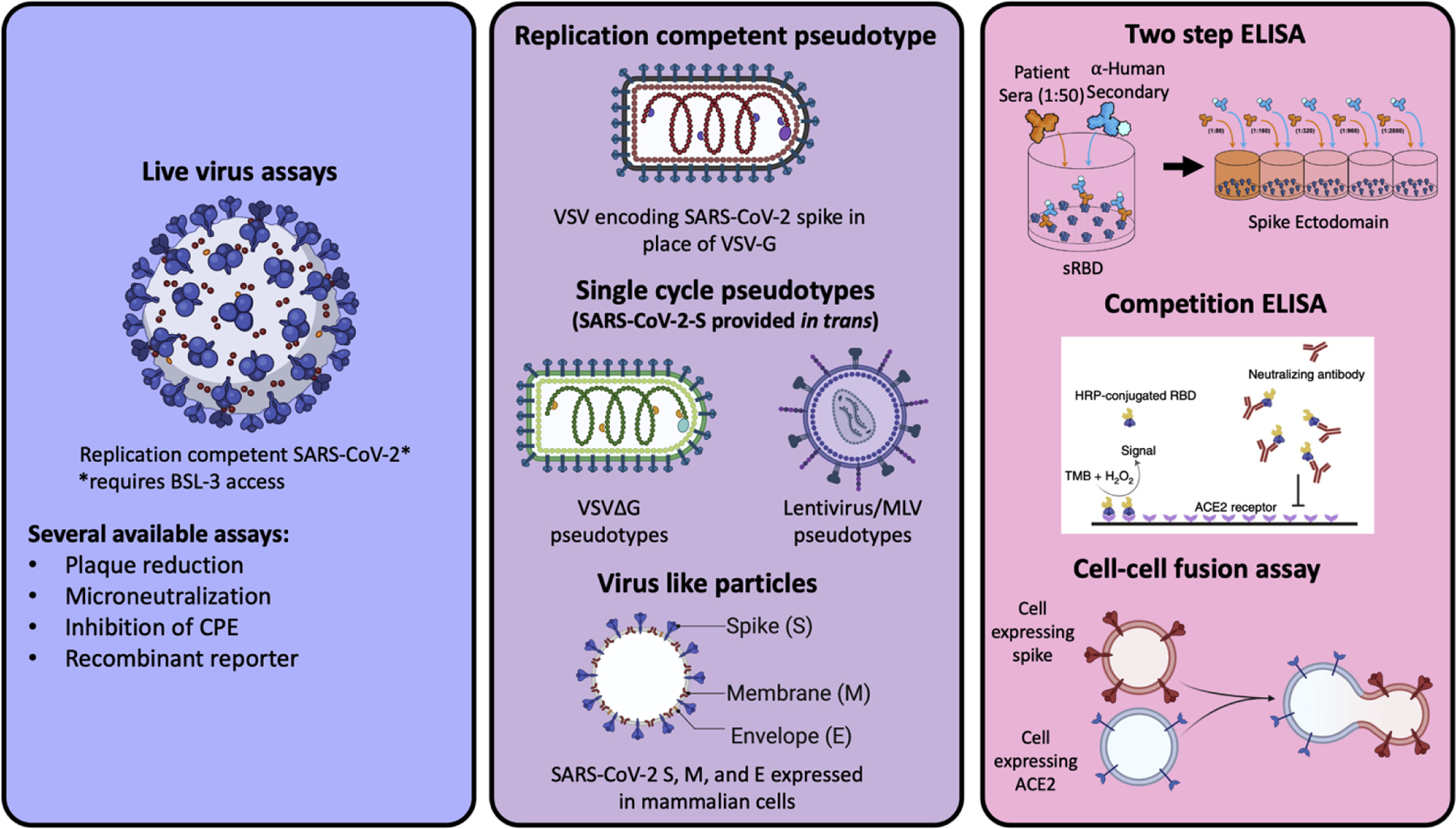
SARS-CoV-2, VSV, lentivirus, virus like particle, and cell-cell fusion assay schematics from Biorender. Two step ELISA schematic generated by Griffin Haas and competition ELISA from Fig 1 of Tan CW et al. [65]
While only a small fraction of papers utilize exogenous proteases or membrane-bound proteases like TMPRSS2 as a part of their entry assays, there can be distinct advantages to utilizing multiple assays in parallel, comparing entry in 293T-ACE2s, highly infectable isogenic 293T-ACE2-TMPRSS2s, and Vero-TMPRSS2s [35,36]. By combining the insight provided from the use of multiple relevant cell lines with the investigation of multiple SARS-CoV-2 spike variants, studies may better resolve the variable efficacy of vaccine sera against variants. Many emerging VOCs/VOIs harbor mutations that might modulate protease priming and entry efficiency. The full picture of antibody protection is not clear without taking into account the variable proteolytic activation of spike that might occur with certain variants under different assay conditions. All in vitro assays of viral entry are inherently imperfect models of human infection, and no one cell line or assay would suffice to recapitulate the myriad cell types and tissues that SARS-CoV-2 can enter and replicate in vivo. Nonetheless, it is important to have conversations about the difficult tradeoffs when designing experiments that match the underlying research questions [37].
Implications
In the case of SARS-CoV-2 variants, the implications of capturing or not capturing the wider picture of relevant proteolytic activation are more immediately obvious. One of the first mutations to become dominant over the earliest Wuhan-Hu-1 genotype was the D614G mutation [38,39] (Fig. 1B). The D614G mutation results in improved stabilization of the spike trimer and increased spike density on the virion [38,40]. In addition, there is evidence that D614G changes S conformation, resulting in better S1/S2 cleavage [41,42] as well as greater neutralization sensitivity. In pseudovirus systems, G614 appears to be more susceptible to neutralization by monoclonal antibodies and hyperimmune patient sera [43]. The same has also been shown in hamsters, with those exposed to D614 having higher neutralization titers against G614 than D614 [44]. The D614G mutation is often discussed within the context of its effect on promoting the accessibility of the receptor binding domain (RBD) [45]; the ‘RBD-up’ conformation potentially exposing vulnerable epitopes for neutralization. However, the data on D614G’s effect on cleavage efficiency also suggests a potential interplay between proteolytic activation and neutralization. Interestingly, the D614G mutation introduces a potential NE cleavage site [46]. NE is a known activator of SARS-CoV infection and can reach extremely high concentrations in the lung during acute infection due to its release by infiltrating neutrophils, potentially exacerbating viral infectivity at the peak of the immune response [31]. Given that alpha-1 antitrypsin is both ubiquitous and a potent inhibitor of NE, the introduction of an elastase cleavage site via D614G is proposed to enhance SARS-CoV-2 spread where alpha-1 antitrypsin concentrations are low [46]. Given the wide array of potential biological differences between D614 and G614 viruses and the fact that G614 is now the prevailing dominant mutation amongst current circulating strains, the continued use of the original D614 viral stocks is not best practice.
There are also now numerous examples of convergent evolution in areas more proximal to the cleavage sites in spike. Position 677 is just outside the furin binding pocket [47] [28] (Fig. 1A/B) and yet multiple lineages have independently developed mutations at the site. Specifically, six independent lineages in the United States have been found containing Q677H and one has been found with Q677P. When modeled, these mutations may lead to a favorable kink that allows higher accessibility to the S1/S2 cleavage site [48].
Another position of interest is at 681, the proline immediately before the furin-like multibasic cleavage site, 681-PRRAR^S-686 (Fig. 1A/B). It is important to note that this multibasic cleavage site is not the classical consensus furin cleavage site, RX[K/R]R^, where dibasic residues at the P1 and P2 positions are preferred. However, substitutions at the P681 to more basic H or R residues might increase the efficiency of cleavage at this S1/S2 junction even if these substitutions do not confer a canonical furin cleavage site [49]. The lineage B.1.617, first identified in India, arose containing P681R [50] and the lineage B.1.1.7, first identified in the U.K., arose containing P681H [49]. P681 mutants are the fastest rising mutants over the first three months of 2021 as measured by the percent of sequences deposited to GISAID containing the mutation of interest [50]. It has been found in the B.1.243 and B.1.222 clades, identified in New York State; A.VOI.V2 found in Angola and Hawaii; and B.1.1.28 identified in the Philippines. MLV-based pseudotypes containing spikes with the P681H substitution appear to be cleaved more efficiently than WT spike. In vitro fluorogenic peptide cleavage assays suggest that P681H renders the S1/S2 junction more susceptible to cleavage by furin at low pH, whereas trypsin mediated cleavage was unaffected [49].
Interestingly, other human beta-coronaviruses do have perfect consensus furin cleavage sites in their spike proteins (RRSRR in HCoV-OC43 and RRKRR in HCoV-HKU1) that are predicted to result in more efficient S1/S2 cleavage [50]. If more efficient furin cleavage per se confers a selective advantage, one might expect selection for a perfect consensus furin cleavage site in SARS-CoV-2. This has not been seen. One hypothesis is that improved cleavability by furin may result in increased cell-cell fusion but not necessarily increased transmissibility and viral fitness. Enhanced fusogenicity appears to compromise viral titers as in the case of SARS-CoV [51,52].
Other mutations of potential importance have been found primarily through in vitro passaging experiments with replication-competent VSV containing the SARS-CoV-2 spike (rVSV-CoV2-S). One mutation appears quite near the S2’ site, P812R (Fig. 1A/B). Its emergence was discovered upon serial passaging and was associated with higher viral titers as well as a lack of syncytia formation [53]. In addition, when rVSV-CoV2-S was used in serial passaging experiments to explore the mutations that form during antibody escape studies, R682Q (PQRAR) was among the most abundant found across nearly all conditions [54] (Fig. 1A/B). Importantly, both of these experiments took place in Vero E6 cells. In the latter study, R682Q was present in their starting inoculum and increased to similarly high frequencies in the absence of any antibody selection, suggesting that the mutation was an adaptation to cell culture conditions. Examples like these illustrate the potential importance of utilizing systems incorporating multiple mechanisms of proteolytic activation. Vero E6 cells do not contain TMPRSS2, nor were other proteases like trypsin or NE integrated into the experimental conditions of the vast majority of studies that use live virus or rVSV-CoV2-S. This may affect what cell culture adaptations form and may also end up affecting the downstream biology of the system in unintended ways.
The choice of cell type may have profound unintended downstream effects. For example, mutations in the multibasic furin-like cleavage site, including large deletions at the S1/S2 site [55,56], commonly occur while passaging viruses in vitro on TMPRSS2-deficient cells such as Vero cells [19]. And losing these sites has a significant impact on the pathogenicity of the virus, whether it be in hamsters [55,56] or other human cell lines [52] where proteases like furin and TMPRSS2 are shown to be vital. Studying SARS-CoV-2 under conditions where the effect of proteases is less relevant can cause one to miss driving factors in variable pathogenicity, such as the impacts of human variation and genetic susceptibility from mutations in TMPRSS2, furin, protease inhibitors like alpha-1 antitrypsin [57] and a large number of others predicted to play a role [58–60].
Loss of S1/S2 cleavage efficiency also has been shown to result in less efficient inhibition by COVID-19 positive patient sera and monoclonal antibodies [61]. Therefore, maintenance of this site is not only important in the biology of proteolytic activation but plays a role in the determination of neutralizing efficacy of antibodies. The latter is underscored by the class of neutralizing antibodies directed against the NTD of the S1 subunit. Some of these antibodies are pH sensitive, and lose their neutralization potential when entry occurs predominantly in the endosomes as in Vero cells (Fig. 1C, late pathway) [62]. SARS-CoV-2 virions without the multibasic cleavage site even show altered sensitivity to interferon and IFITM2 [55]. All of this occurs, in part, because S1/S2 cleavage is necessary for S2’ cleavage at the cell surface via TMPRSS2 in Calu-3, but not for S2’ activation via cathepsin L in Vero E6 [18,58].
It is now clear that loss of the WT furin-like cleavage site occurs easily in cell culture when viruses are passaged on TMPRSS2-deficient cells, and that maintenance of this WT cleavage site is important for evaluating the neutralizing activities of antibodies. Alarmingly, the majority of SARS-CoV-2 isolates currently available from BEI resources have one or more mutations or deletions that affect the furin cleavage site. Table 2 lists the SARS-CoV-2 isolates from BEI resources and their associated NGS data which pinpoint the cognate furin site mutations. We urge the field to be cognizant of these potential confounding factors. The use of TMPRSS2-expressing cell lines like Calu-3 or Vero-TMPRSS2, incorporating trypsin use, or performing experiments in relevant organoids [19,63] are all possible solutions to maintaining WT cleavage site sequences when culturing out viruses and/or expanding viral stocks. Our current review has shown that the biology of SARS-CoV-2 entry is highly dependent on the interplay between cleavage site mutations and the proteases present. It is vital to be cognizant of both these factors, especially when assessing patient sera from myriad vaccines against multiple variants or when screening for monoclonal antibody efficacy where one cannot rule out a priori the role that different proteases may play in the entry and replication of a given virus isolate [61].
Table 2. SARS-CoV-2 isolates from BEI resources carrying mutations near the S1/S2 cleavage site.
All SARS-CoV-2 isolates available as of July 6th, 2021 from BEI resources that contain mutations near the S1/S2 cleavage site. Data was compiled using the certificates of analysis provided by BEI resources. Also included is a recombinant infectious clone derived from USA-WA1/2020 isolate found to contain two mutations in the S1/S2 cleavage site. Yellow cells indicate substitutions and red cells indicate deletions. Not included are the 17 other available isolates containing either no mutations at S1/S2 or containing mutations canonical to their lineage (e.g. P681R in B.1.617 or P681H in B.1.17). Mutations noted are relative to MN908947.3, the Wuhan-Hu-1 SARS-CoV-2 isolate.
| BEI Number | Name (SARS-CoV-2 isolate) | Lineage | Native Furin Site AA Seq | Frequency |
|---|---|---|---|---|
| 675- QTQTNSPRRAR^S −686 | ||||
| NR-53945 | Isolate hCoV-19/Scotland/CVR2224/2020 | B.1.222 | Mut: R682L | 0.060 |
| NR-52368 | Isolate New York 1-PV08001/2020 | B.4 | Mut: R682W | 0.402 |
| NR-52281 | Isolate USA-WA1/2020 | A | Mut: R682L | 0.159 |
| NR-52282 | Isolate Hong Kong/VM20001061/2020 | A | Mut: R682Q | 0.958 |
| NR-52386 | Isolate USA-CA4/2020 | B | Mut: R682L | 0.817 |
| NR-52387 | Isolate USA-CA2/2020 | B.2 | Mut: R682L | 0.587 |
| NR-52439 | Isolate Chile/Santiago_op4d1/2020 | A.2 | Mut: R685H | 0.989 |
| NR-53514 | Isolate New York-PV08410/2020 | B.1 | Mut: R682W | 0.439 |
| NR-53516 | Isolate New York-PV09158/2020 | B.1.3 | Mut: R682Q | 0.583 |
| NR-53565 | Isolate Canada/ON/VIDO-01/2020 | B | Mut: R685S | 0.463 |
| NR-54009 | Isolate hCoV-19/South Africa/KRISPK005325/2020 | B.1.351 | Mut: R682W | 0.902 |
| Mut: Q677H | 0.899 | |||
| NR-52284 | SARS-CoV-2, Isolate Italy-INMI1 | Not Stated | Mut: R682W | 0.209 |
| 679–686 del | 0.611 | |||
| NR-52359 | SARS-CoV-2, Isolate England/02/2020 | A | 679–686 del | 0.824 |
| NR-52370 | SARS-CoV-2, Isolate Germany/BavPat1/2020 | B | 679–684 del | 0.565 |
| NR-52381 | SARS-CoV-2, Isolate USA-IL1/2020 | B | 675–679 del | 0.613 |
| NR-52382 | SARS-CoV-2, Isolate USA-CA1/2020 | A | 675–679 del | 0.613 |
| NR-52383 | SARS-CoV-2, Isolate USA-AZ1/2020 | A | 681–687 del | 0.861 |
| NR-52384 | SARS-CoV-2, Isolate USA-WI1/2020 | B | 687–689 del | 0.527 |
| BEI Number | Name | Lineage | Result | Frequency |
| NR-54003 | SARS-CoV-2, Isolate USA-WA1/2020, Recombinant Infectious Clone with Nanoluciferase Gene (icSARS-CoV-2-nLuc) | A | A684E | 0.287 |
| A | S686G | 0.104 |
Conclusions
Proteolytic priming, activation, and receptor binding are critical modulators of SARS-CoV-2 entry. Multiple non-exclusive entry pathways that are dependent on the unique contexts of SARS-CoV-2 sequence variability, the constellation of local proteases, and their regulators such as alpha-1 antitrypsin highlights the complexity of assessing SARS-CoV-2 entry.
There are significant implications of this complexity on entry assays that will drive our understanding of humoral immunity, therapeutic efficacy of entry inhibitors, and tissue restriction. Few studies looking at therapeutics like vaccines and convalescent plasma measure entry under multiple conditions; comparing different proteases, protease inhibitors, and spike sequences. Many utilize cell lines, cell-cultured adapted viruses with no sequence verification, and pseudovirus sequences that are known to have unintended effects on cleavage efficiency, entry kinetics, and antibody efficacy.
We call for improved standards including 1) that entry inhibition assays include conditions that utilize cells containing TMPRSS2 and/or include the use of proteases like trypsin, 2) that virus or pseudovirus rescue and passaging occur on cells containing TMPRSS2 and/or in the presence of proteases like trypsin, 3) that therapeutic efficacy be tested against multiple relevant SARS-CoV-2 spike variants of interest and/or dominant variants at the time, especially with respect to those with known effects on spike cleavage; whether with live virus, replication-competent pseudovirus, or replication-incompetent pseudoviruses.
In Castillo-Olivares et al [64], they work to identify universal standards for correlate of protection and importantly use 293T cells expressing both ACE2 and TMPRSS2 when performing their SARS-CoV-2 pseudotype-based microneutralization assay and used Vero-ACE2/TMPRSS2 cells for their electroporation-dependent neutralization assay and their intracellular neutralization assay. We argue that the use of increasingly relevant cell types and protease contexts could be especially powerful in concert with spike variants of interest for identifying universal standards for correlates of protection and also for providing better grounds on which to compare data generally across the ever-increasing body of SARS-CoV-2 literature.
Acknowledgements
We thank members of the Lee lab for discussions and input. KYO and CSS acknowledge support from the Training Program in Mechanisms of Virus-Host Interactions (T32 AI07647). KYO is additionally supported by NRSA F31 predoctoral fellowship AI154739. BL acknowledges support from the Department of Microbiology at the Icahn School of Medicine, and re-directed support from NIH grants AI123449, AI125536, AI071002, AI138921, AI115226.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none
References
- 1.Perlman S: Another decade, another Coronavirus. N Engl J Med 2020, 382:760–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, et al. : A novel Coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020, 382:727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lever J, Altman RB: Analyzing the vast coronavirus literature with CoronaCentral. Proc Natl Acad Sci U S A 2021, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, Graham BS, McLellan JS: Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367:1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY, et al. : Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181:894–904.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Letko M, Marzi A, Munster V: Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nature Microbiology 2020, 5:562–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang T, Bidon M, Jaimes JA, Whittaker GR, Daniel S: Coronavirus membrane fusion mechanism offers as a potential target for antiviral development. Antiviral Res 2020, 178:104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann M, Kleine-Weber H, Schroeder S, Mü MA, Drosten C, Pö S, Krü N, Herrler T, Erichsen S, Schiergens TS, et al. : SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor Article SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** One of the first comprehensive characterization of SARS-CoV-2 entry and tropism.
- 9.Bestle D, Heindl MR, Limburg H, Van Lam van T, Pilgram O, Moulton H, Stein DA, Hardes K, Eickmann M, Dolnik O, et al. : TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci Alliance 2020, 3:e202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Careful analysis of proteolytic activation processes that regulate SARS-CoV-2 entry into physiologically relevant primary airway cells.
- 10.Cai Y, Zhang J, Xiao T, Peng H, Sterling SM, Walsh RM Jr, Rawson S, Rits-Volloch S, Chen B: Distinct conformational states of SARS-CoV-2 spike protein. Science 2020, 369:1586–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whittaker GR, Daniel S, Millet JK: Coronavirus entry: how we arrived at SARSCoV-2. Curr Opin Virol 2021, 47:113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitra P: Inhibiting fusion with cellular membrane system: therapeutic options to prevent severe acute respiratory syndrome coronavirus-2 infection. Am J Physiol Cell Physiol 2020, 319:C500–C509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papa G, Mallery DL, Albecka A, Welch LG, Cattin-Ortolá J, Luptak J, Paul D, McMahon HT, Goodfellow IG, Carter A, et al. : Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell-cell fusion. PLoS Pathog 2021, 17:e1009246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann M, Kleine-Weber H, Pöhlmann S: A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell 2020, 78:779–784.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winstone H, Lista MJ, Reid AC, Bouton C, Pickering S, Galao RP, Kerridge C, Doores KJ, Swanson CM, Neil SJD: The polybasic cleavage site in SARS-CoV-2 spike modulates viral sensitivity to type I interferon and IFITM2. J Virol 2021, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E: The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res 2020, 176:104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang T, Jaimes JA, Bidon MK, Straus MR, Daniel S, Whittaker GR: Proteolytic activation of SARS-CoV-2 spike at the S1/S2 boundary: Potential role of proteases beyond furin. ACS Infect Dis 2021, 7:264–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bollavaram K, Leeman TH, Lee MW, Kulkarni A, Upshaw SG, Yang J, Song H, Platt MO: Multiple sites on SARS-CoV-2 spike protein are susceptible to proteolysis by cathepsins B, K, L, S, and V. Protein Sci 2021, 30:1131–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sasaki M, Uemura K, Sato A, Toba S, Sanaki T, Maenaka K, Hall WW, Orba Y, Sawa H: SARS-CoV-2 variants with mutations at the S1/S2 cleavage site are generated in vitro during propagation in TMPRSS2-deficient cells. PLoS Pathog 2021, 17:e1009233. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Well-documented experiments showing the easy accumulation of cleavage site mutations during cell culture passage in TMPRSS2-negative cells.
- 20.Simmons G, Zmora P, Gierer S, Heurich A, Pöhlmann S: Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res 2013, 100:605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mustafa Z, Zhanapiya A, Kalbacher H, Burster T: Neutrophil Elastase and Proteinase 3 Cleavage Sites Are Adjacent to the Polybasic Sequence within the Proteolytic Sensitive Activation Loop of the SARS-CoV-2 Spike Protein. ACS Omega 2021, 6:7181–7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wettstein L, Weil T, Conzelmann C, Müller JA, Groß R, Hirschenberger M, Seidel A, Klute S, Zech F, Prelli Bozzo C, et al. : Alpha-1 antitrypsin inhibits TMPRSS2 protease activity and SARS-CoV-2 infection. Nat Commun 2021, 12:1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hantak MP, Qing E, Earnest JT, Gallagher T: Tetraspanins: Architects of viral entry and exit platforms. J Virol 2019, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohamed MMA, El-Shimy IA, Hadi MA: Neutrophil Elastase Inhibitors: A potential prophylactic treatment option for SARS-CoV-2-induced respiratory complications? Crit Care 2020, 24:311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chambers JP, Yu J, Valdes JJ, Arulanandam BP: SARS-CoV-2, early entry events. J Pathog 2020, 2020:9238696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peacock TP, Goldhill DH, Zhou J, Baillon L, Frise R, Swann OC, Kugathasan R, Penn R, Brown JC, Sanchez-David RY, et al. : The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol 2021, doi: 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]; * Important paper showing the importance of the intact furin site for transmissibility.
- 27.Savela ES, Winnett A, Romano AE, Porter MK, Shelby N, Akana R, Ji J, Cooper MM, Schlenker NW, Reyes JA, et al. : SARS-CoV-2 is detectable using sensitive RNA saliva testing days before viral load reaches detection range of low-sensitivity nasal swab tests. medRxiv 2021, doi: 10.1101/2021.04.02.21254771. [DOI] [Google Scholar]
- 28.Huang N, Pérez P, Kato T, Mikami Y, Okuda K, Gilmore RC, Conde CD, Gasmi B, Stein S, Beach M, et al. : SARS-CoV-2 infection of the oral cavity and saliva. Nat Med 2021, 27:892–903. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Important study showing the variety of proteases in physiologically relevant locations that can prime and activate SARS-CoV-2 spike.
- 29.Klingler J, Lambert GS, Itri V, Liu S, Bandres JC, Enyindah-Asonye G, Liu X, Oguntuyo KY, Amanat F, Lee B, et al. : SARS-CoV-2 mRNA vaccines induce a greater array of spike-specific antibody isotypes with more potent complement binding capacity than natural infection. medRxiv 2021, doi: 10.1101/2021.05.11.21256972. [DOI] [Google Scholar]
- 30.Singh M, Bansal V, Feschotte C: A Single-Cell RNA Expression Map of Human Coronavirus Entry Factors. Cell Rep 2020, 32:108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuyama S, Ujike M, Morikawa S, Tashiro M, Taguchi F: Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc Natl Acad Sci U S A 2005, 102:12543–12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuentes-Prior P: Priming of SARS-CoV-2 S protein by several membrane-bound serine proteinases could explain enhanced viral infectivity and systemic COVID-19 infection. J Biol Chem 2020, 296:100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simonovich VA, Burgos Pratx LD, Scibona P, Beruto MV, Vallone MG, Vázquez C, Savoy N, Giunta DH, Pérez LG, Sánchez MDL, et al. : A Randomized Trial of Convalescent Plasma in Covid-19 Severe Pneumonia. N Engl J Med 2021, 384:619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perotti C, Baldanti F, Bruno R, Del Fante C, Seminari E, Casari S, Percivalle E, Glingani C, Musella V, Belliato M, et al. : Mortality reduction in 46 severe Covid-19 patients treated with hyperimmune plasma. A proof of concept single arm multicenter trial. Haematologica 2020, 105:2834–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oguntuyo KY, Stevens CS, Hung CT, Ikegame S, Acklin JA, Kowdle SS, Carmichael JC, Chiu H-P, Azarm KD, Haas GD, et al. : Quantifying Absolute Neutralization Titers against SARS-CoV-2 by a Standardized Virus Neutralization Assay Allows for Cross-Cohort Comparisons of COVID-19 Sera. MBio 2021, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Comprehensive analysis of parameters that govern efficient SARS-CoV-2 entry.
- 36.Ikegame S, Siddiquey MNA, Hung C-T, Haas G, Brambilla L, Oguntuyo KY, Kowdle S, Vilardo AE, Edelstein A, Perandones C, et al. : Qualitatively distinct modes of Sputnik V vaccine-neutralization escape by SARS-CoV-2 Spike variants. medRxiv 2021, doi: 10.1101/2021.03.31.21254660. [DOI] [Google Scholar]
- 37.Khoury DS, Wheatley AK, Ramuta MD, Reynaldi A, Cromer D, Subbarao K, O’Connor DH, Kent SJ, Davenport MP: Measuring immunity to SARS-CoV-2 infection: comparing assays and animal models. Nat Rev Immunol 2020, 20:727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L, Jackson CB, Mou H, Ojha A, Peng H, Quinlan BD, Rangarajan ES, Pan A, Vanderheiden A, Suthar MS, et al. : SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun 2020, 11:6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, et al. : Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182:812–827.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Seminal study. First one to call out and characterize the D614G variant that is now the dominant background for all SARS-CoV-2 spike.
- 40.Zhang J, Cai Y, Xiao T, Lu J, Peng H, Sterling SM, Walsh RM Jr, Rits-Volloch S, Zhu H, Woosley AN, et al. : Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science 2021, 372:525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gobeil SM-C, Janowska K, McDowell S, Mansouri K, Parks R, Manne K, Stalls V, Kopp MF, Henderson R, Edwards RJ, et al. : D614G mutation alters SARS-CoV-2 spike conformation and enhances protease cleavage at the S1/S2 junction. Cell Rep 2021, 34:108630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daniloski Z, Jordan TX, Ilmain JK, Guo X, Bhabha G, tenOever BR, Sanjana NE: The Spike D614G mutation increases SARS-CoV-2 infection of multiple human cell types. Elife 2021, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weissman D, Alameh M-G, de Silva T, Collini P, Hornsby H, Brown R, LaBranche CC, Edwards RJ, Sutherland L, Santra S, et al. : D614G Spike Mutation Increases SARS CoV-2 Susceptibility to Neutralization. Cell Host Microbe 2021, 29:23–31.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, Zhang X, Muruato AE, Zou J, Fontes-Garfias CR, et al. : Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021, 592:116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mansbach RA, Chakraborty S, Nguyen K, Montefiori DC, Korber B, Gnanakaran S: The SARS-CoV-2 Spike variant D614G favors an open conformational state. Sci Adv 2021, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhattacharyya C, Das C, Ghosh A, Singh AK, Mukherjee S, Majumder PP, Basu A, Biswas NK: SARS-CoV-2 mutation 614G creates an elastase cleavage site enhancing its spread in high AAT-deficient regions. Infect Genet Evol 2021, 90:104760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian S, Huajun W, Wu J: Computational prediction of furin cleavage sites by a hybrid method and understanding mechanism underlying diseases. Sci Rep 2012, 2:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hodcroft EB, Domman DB, Snyder DJ, Oguntuyo K, Van Diest M, Densmore KH, Schwalm KC, Femling J, Carroll JL, Scott RS, et al. : Emergence in late 2020 of multiple lineages of SARS-CoV-2 Spike protein variants affecting amino acid position 677. medRxiv 2021, doi: 10.1101/2021.02.12.21251658. [DOI] [Google Scholar]
- 49.Lubinski B, Tang T, Daniel S, Jaimes JA, Whittaker GR: Functional evaluation of proteolytic activation for the SARS-CoV-2 variant B.1.1.7: role of the P681H mutation. bioRxivorg 2021, doi: 10.1101/2021.04.06.438731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cherian S, Potdar V, Jadhav S, Yadav P, Gupta N, Das M, Rakshit P, Singh S, Abraham P, Panda S, et al. : Convergent evolution of SARS-CoV-2 spike mutations, L452R, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. 2021, doi: 10.1101/2021.04.22.440932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Belouzard S, Chu VC, Whittaker GR: Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci U S A 2009, 106:5871–5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Follis KE, York J, Nunberg JH: Furin cleavage of the SARS coronavirus spike glycoprotein enhances cell–cell fusion but does not affect virion entry. Virology 2006, 350:358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dieterle ME, Haslwanter D, Bortz RH 3rd, Wirchnianski AS, Lasso G, Vergnolle O, Abbasi SA, Fels JM, Laudermilch E, Florez C, et al. : A Replication-Competent Vesicular Stomatitis Virus for Studies of SARS-CoV-2 Spike-Mediated Cell Entry and Its Inhibition. Cell Host Microbe 2020, 28:486–496.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baum A, Fulton BO, Wloga E, Copin R, Pascal KE, Russo V, Giordano S, Lanza K, Negron N, Ni M, et al. : Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 2020, 369:1014–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lau S-Y, Wang P, Mok BW-Y, Zhang AJ, Chu H, Lee AC-Y, Deng S, Chen P, Chan K-H, Song W, et al. : Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg Microbes Infect 2020, 9:837–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hale BG: Avoiding culture shock with the SARS-CoV-2 spike protein. Elife 2021, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oguntuyo KY, Stevens CS, Siddiquey MN, Schilke RM, Woolard MD, Zhang H, Acklin JA, Ikegame S, Hung C-T, Lim JK, et al. : In plain sight: the role of alpha-1-antitrypsin in COVID-19 pathogenesis and therapeutics. bioRxiv 2020, [Google Scholar]
- 58.Klaassen K, Stankovic B, Zukic B, Kotur N, Gasic V, Pavlovic S, Stojiljkovic M: Functional prediction and comparative population analysis of variants in genes for proteases and innate immunity related to SARS-CoV-2 infection. Infect Genet Evol 2020, 84:104498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laporte M, Raeymaekers V, Van Berwaer R, Vandeput J, Marchand-Casas I, Thibaut H-J, Van Looveren D, Martens K, Hoffmann M, Maes P, et al. : The SARS-CoV-2 and other human coronavirus spike proteins are fine-tuned towards temperature and proteases of the human airways. PLoS Pathog 2021, 17:e1009500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Torre-Fuentes L, Matías-Guiu J, Hernández-Lorenzo L, Montero-Escribano P, Pytel V, Porta-Etessam J, Gómez-Pinedo U, Matías-Guiu JA: ACE2, TMPRSS2, and Furin variants and SARS-CoV-2 infection in Madrid, Spain. J Med Virol 2021, 93:863–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnson BA, Xie X, Bailey AL, Kalveram B, Lokugamage KG, Muruato A, Zou J, Zhang X, Juelich T, Smith JK, et al. : Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 2021, 591:293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Shows that furin site in live SARS-CoV-2 is important for pathogenesis in vivo and also important for maintaining full sensitivity to neutralizing antibodies.
- 62.McCallum M, De Marco A, Lempp FA, Tortorici MA, Pinto D, Walls AC, Beltramello M, Chen A, Liu Z, Zatta F, et al. : N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 2021, 184:2332–2347.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lamers MM, Mykytyn AZ, Breugem TI, Wang Y, Wu DC, Riesebosch S, van den Doel PB, Schipper D, Bestebroer T, Wu NC, et al. : Human airway cells prevent SARS-CoV-2 multibasic cleavage site cell culture adaptation. Elife 2021, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Castillo-Olivares J, Wells DA, Ferrari M, Chan A, Smith P, Nadesalingam A, Paloniemi M, Carnell G, Ohlendorf L, Cantoni D, et al. : Towards Internationally standardised humoral Immune Correlates of Protection from SARS-CoV-2 infection and COVID-19 disease. 2021, doi: 10.1101/2021.05.21.21257572. [DOI] [Google Scholar]
- 65.Tan CW, Chia WN, Qin X, Liu P, Chen MI-C, Tiu C, Hu Z, Chen VC-W, Young BE, Sia WR, et al. : A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2–spike protein–protein interaction. Nat Biotechnol 2020, 38:1073–1078. [DOI] [PubMed] [Google Scholar]


