Figure 2.
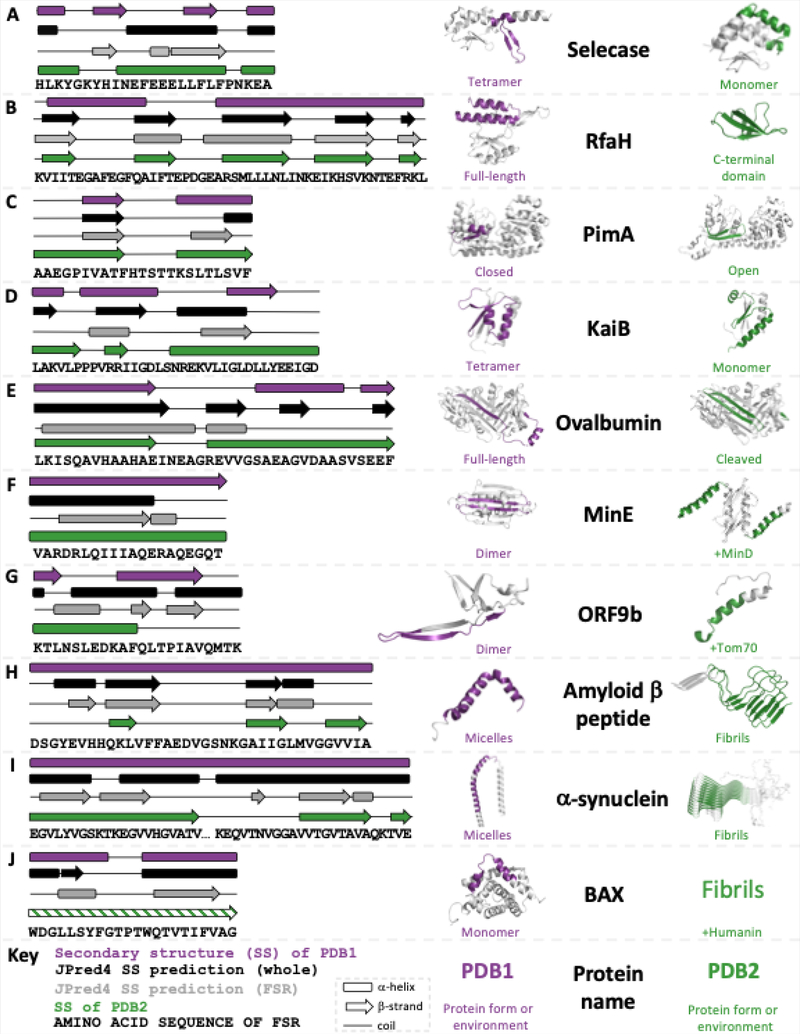
JPred4 predicts different secondary structures for isolated and contextualized FSRs of extant fold switchers with substantive transitions between α-helix and β-strand. Each panel shows the experimentally determined secondary structures of both conformations of the fold switcher (purple and green) along with JPred4 secondary structure predictions of the whole sequence (black) and FSR (gray). Purple and green regions of protein structure correspond to FSR sequence shown in diagram; gray correspond to Structurally Constant Regions (SCRs). Predicted secondary structures that were at least 2 contiguous residues long are shown. The KaiB variant (2QKE) represents all members of the KaiB family; the other 3 (1WWJ, 4SKO, 1R5P) are not shown; amylin is also not shown due to lack of space. The differential secondary structure predictions for ORF9b were reported previously40. The green secondary structure diagram of BAX is shaded with lines to signify that its structure has not been solved, though other experimental evidence strongly suggests that it folds into a β-sheet. All three-dimensional protein structures were made using PyMOL.