Fig. 4. A four-residue loop in coronavirus nsp16 alters the orientation of the RNA substrate.
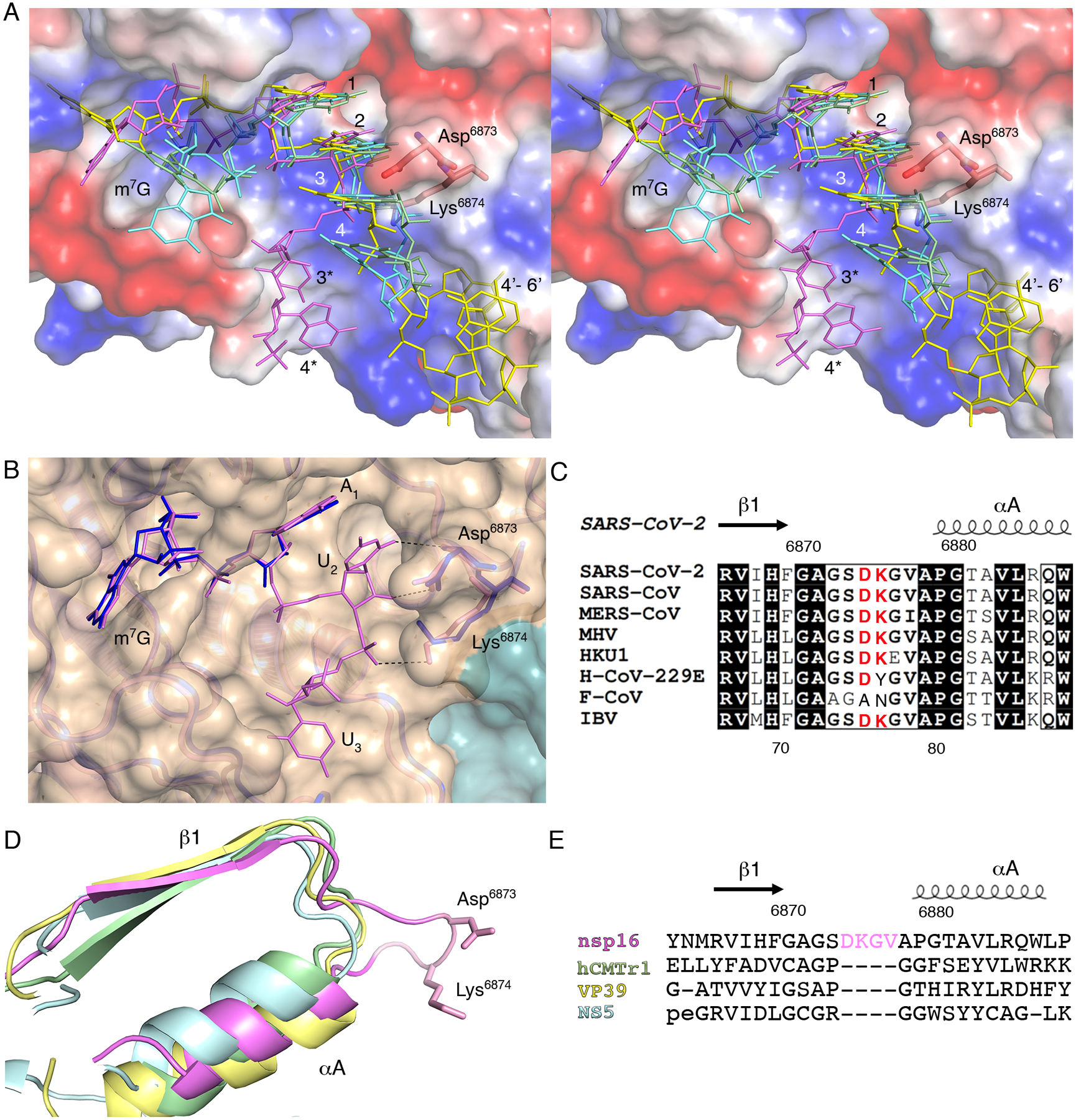
(A) Wall-eyed stereo view of the superimposed capped RNAs mapped on the electrostatic potential surface of SARS-CoV-2 2′-O-MTase. RNAs are shown as sticks in bright pink (SARS-CoV-2), cyan (DENV NS5), yellow (VACV VP39) and green (hCMTr1). The nucleotides are labeled with numbers starting from m7G (position 0); 3* and 4* correspond to U3 and A4 in the Cap-1-RNA from SARS-CoV-2, 4’-6’ to A4-A5-A6 in the capped mRNA from VACV. (B) Superimposition of the RNA binding sites of nsp16-nsp10 with Cap-0-RNA (SARS-CoV-2, magenta) and Cap-0 analog (MERS-CoV, blue). Protein chains are shown as ribbons in pink for SARS-CoV-2 (PDB 7JYY) and blue for MERS-CoV (PDB 5YNM) overlayed on the semitransparent solvent-exposed surface of nsp16 (beige) and nsp10 (teal). Direct hydrogen bond interactions of Asp6873 and Lys6874 with U2 are shown as black, dashed lines. (C) Multiple sequence alignment of the loop region between β1 and αA from different coronaviruses, with Asp6873 and Lys6874 highlighted in red. SARS-CoV, severe acute respiratory syndrome; MERS-CoV, Middle East respiratory syndrome; MHV, murine hepatitis virus (MHV); HKU1, human coronavirus 1; H-CoV-229E, human coronavirus 229E; F-CoV, feline coronavirus; IBV, infectious bronchitis virus (IBV). (D and E) Structural and sequence alignments of nsp16 in pink, hCMTr1 in green, VP39 in yellow, and NS5 in cyan. Protein chains are shown as cartoon models with conserved Asp6873 and Lys6874 from nsp16 shown as sticks and the insertion loop highlighted in pink.
