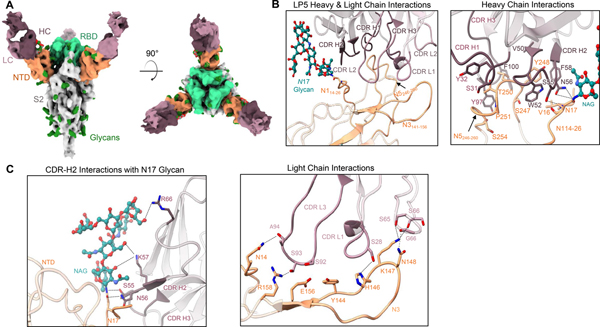
Figure 5. Structural analysis of the NTD-directed neutralizing antibody LP5.
(A) Cryo-EM reconstructions for SARS-CoV-2 spike complexes with LP5. The NTD is shown in orange, RBD in green, glycans in dark green, with the LP5 antibody heavy chains in magenta and light chains in gray. (B) Expanded view of LP5 interactions with NTD showing the overall interface (left), and recognition by the LP5 heavy chain (right), (C) CDR-H2 recognition of the SARS-CoV-2 N17 glycan (left), and recognition of NTD by the LP5 light chain (right). NTD regions N1 (14–26), N3 (residues 141–156) and N5 (residues 246–260) are shown. All hydrogen bonds (distance ~3.2 Å) are represented as dashed lines.