FIGURE 2.
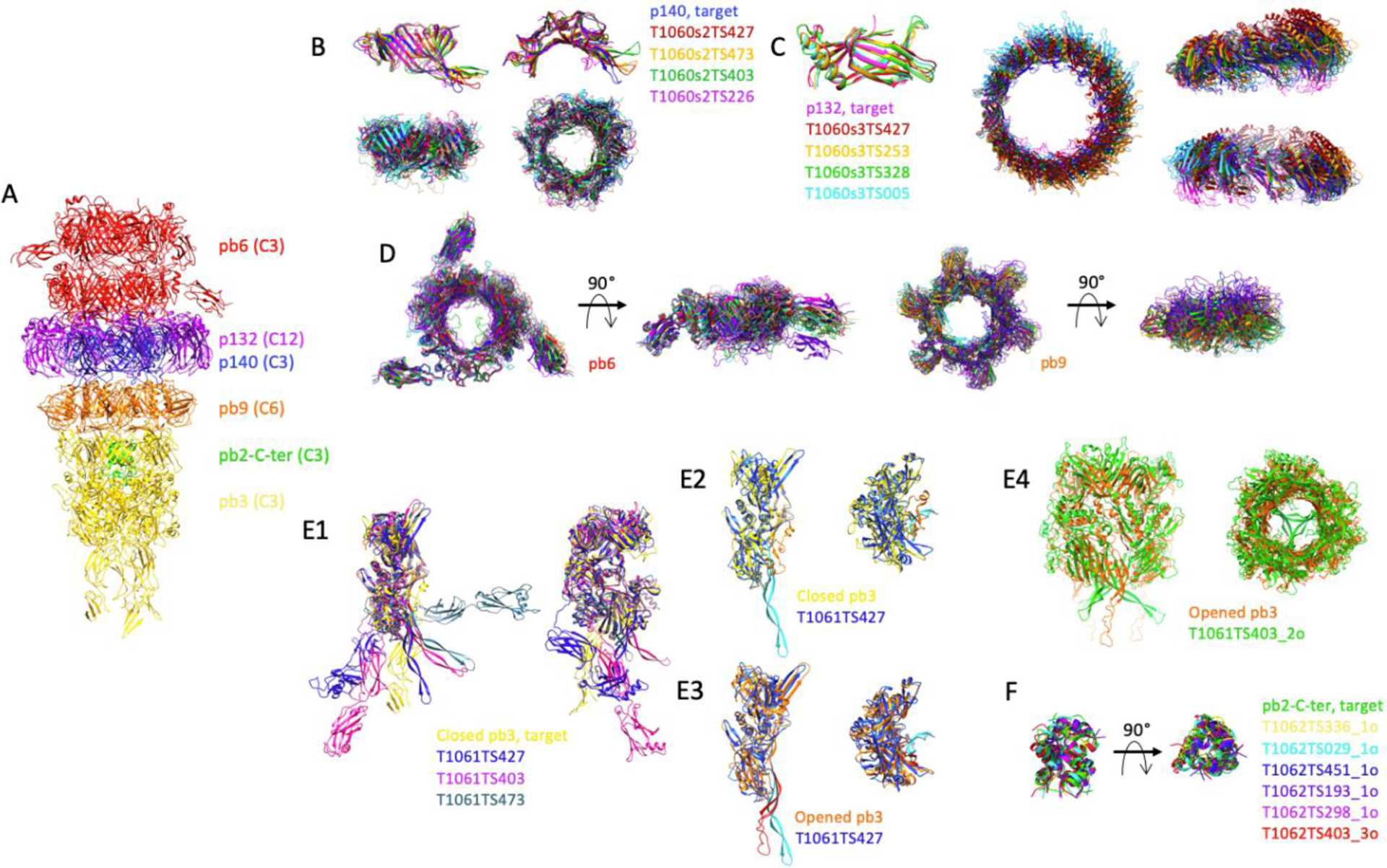
Comparison of the predicted structures of individual proteins and sub-complexes thereof, of the tip of phage T5. A, Experimental structure of phage T5 tail tip. The two pb6 rings are coloured red, that of p140: blue, the p132 dodecamer: purple, the pb9 hexamer: orange, the pb3 trimer: yellow and the pb2-C-terminus: green. The symmetry of each protein is indicated in brackets. B, Monomer (top) and trimer (bottom) predictions for p140. Side (left) and top (right) views. C, Monomer (left) and dodecamer (right) predictions for p132. The dodecamer is shown in a top view and two side views, rotated by 90°. D, Ring prediction for pb6 trimer (left) and pb9 hexamer (right). Top and side views. E1, Monomer predictions for pb3, two side views, rotated by 90°. E2–3, T1061TS427 prediction was superimposed either on the closed pb3 (E2) or on the open pb3 (E3). The 35-residue plug is coloured orange in the closed pb3 and red in the open one, and cyan in the T1061TS427 prediction. Side (left) and top (right) views. E4, Trimer prediction of pb3. The open form of pb3 (orange) is shown rather than the closed one. In E2–4, the fibronectin domains have been removed for clarity. F, trimeric predictions for pb2. In each panel, predicted structures are superimposed on the target, shown in the same colour as in panel A. Superimpositions are done with the Chimera tool MatchMaker. In the case of a multimer, the structures are aligned to one monomer.
