FIGURE 6.
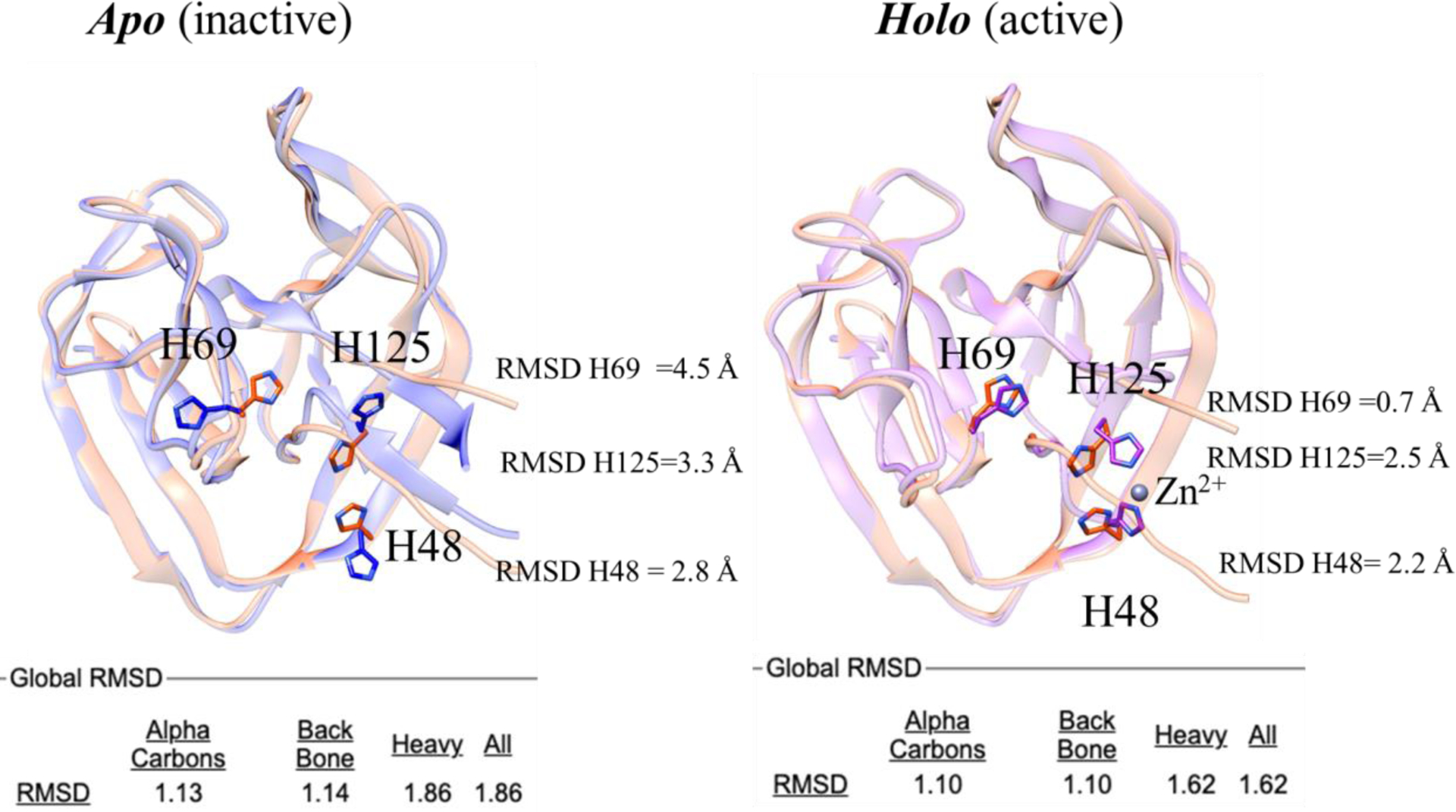
Structure superposition of the 3D model from AlphaFold2 (salmon) with the BIL2 experimentally-determined structures (cornflower blue and magenta). The catalytic residue H69, as well as the Zn coordinating residues H125 and H148 residues, are highlighted in sticks. Labels indicate per-residue RMSD values over backbone and side chains (calculated with CHIMERA). Global RMSD values (calculated with Superpose) over alpha carbons, backbone and side chains atoms are reported in the bottom part of the figure.
