Figure 3. ATG7 and ATG12 are the key targets of NURR1.
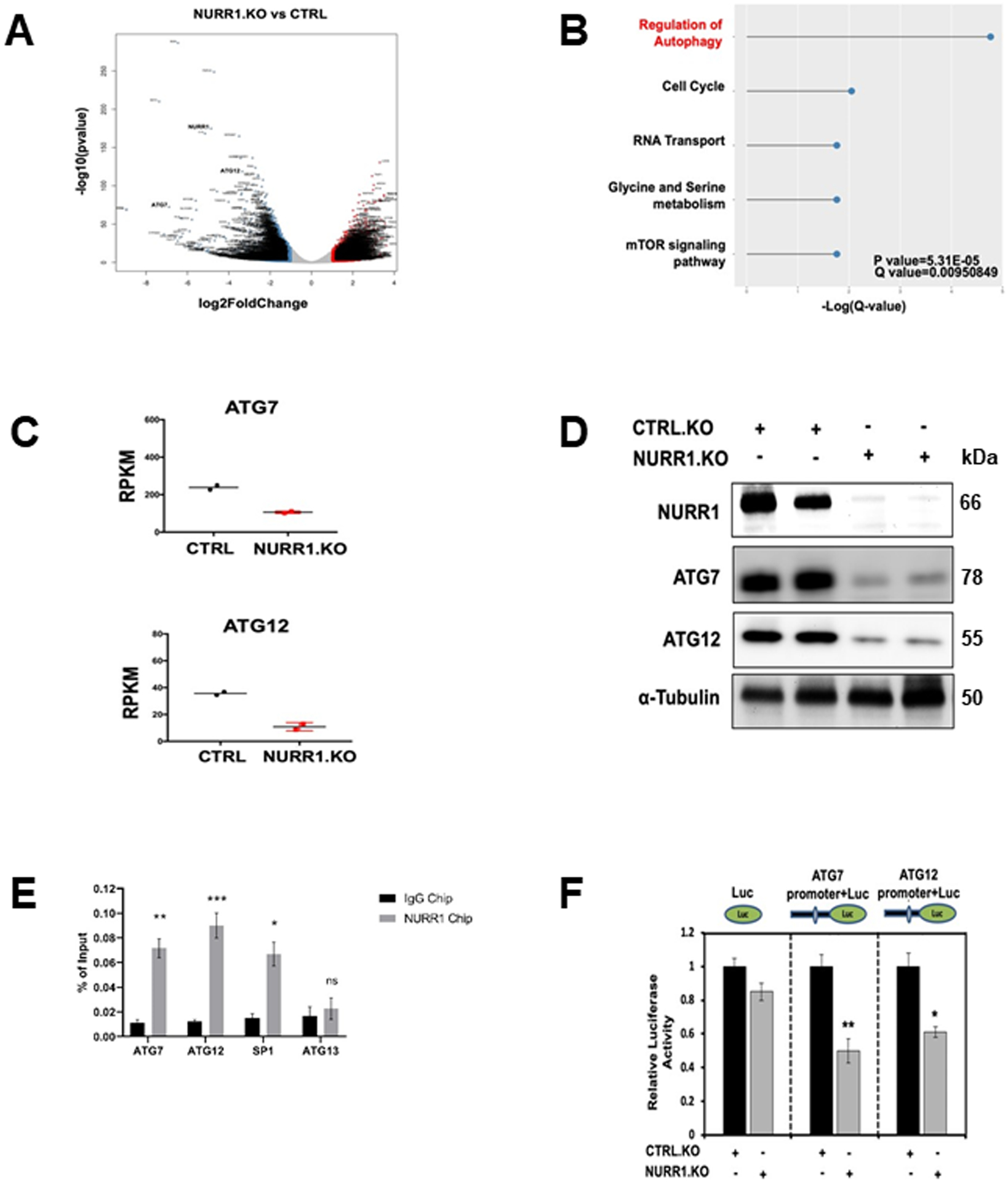
A) Volcano plots of log2fold change vs. −log10(P value) of RNA-Seq data for NURR1.KO vs. control (CTRL), for MiaPaCa2 cells subject to these manipulations. B) KEGG enrichment pathway analysis of NURR1 regulated pathways by comparing gene expressed wild-type MiaPaca2 cells and NRR1 knockout cells. C) RT-qPCR analysis of ATG7and ATG12 expression in CTRL and NURR1.KO cells. Gene expression is normalized to β-actin. Each data point represents the mean ± SEM of four independent experiments (** p < 0.01; *** p < 0.001), D) Immunoblot analysis of NURR1, ATG7 and ATG12 in NURR1.KO cells compared to CTRL MiaPaCa2 cells with α-Tubulin as loading control. E) Q-PCR analysis of a ChIP assay in MiaPaCa2 cells shows a significant increase of NURR1 interactions on the in ATG7 and ATG12 promoters relative to IgG, ** p < 0.01, *** p < 0.001. Results are expressed as the fold enrichment over input DNA. Error bars represent the mean ± SEM of three independent experiments F) Dual luciferase assay of MiaPaCa2 cells with empty vector control (EV) and NURR1.KO. Cell lines were transfected with Renilla luciferase reporter constructs fused with ATG7 or ATG12 promoter, as well as a constitutive firefly luciferase expression construct for 24 h. Renilla luciferase activity was normalized to firefly luciferase activity, and results shown are the average of 4 experiments ± SEM * p < 0.05, ** p < 0.01.
