Figure 4. EP300 degradation rapidly disrupts MYCN expression and causes apoptosis.
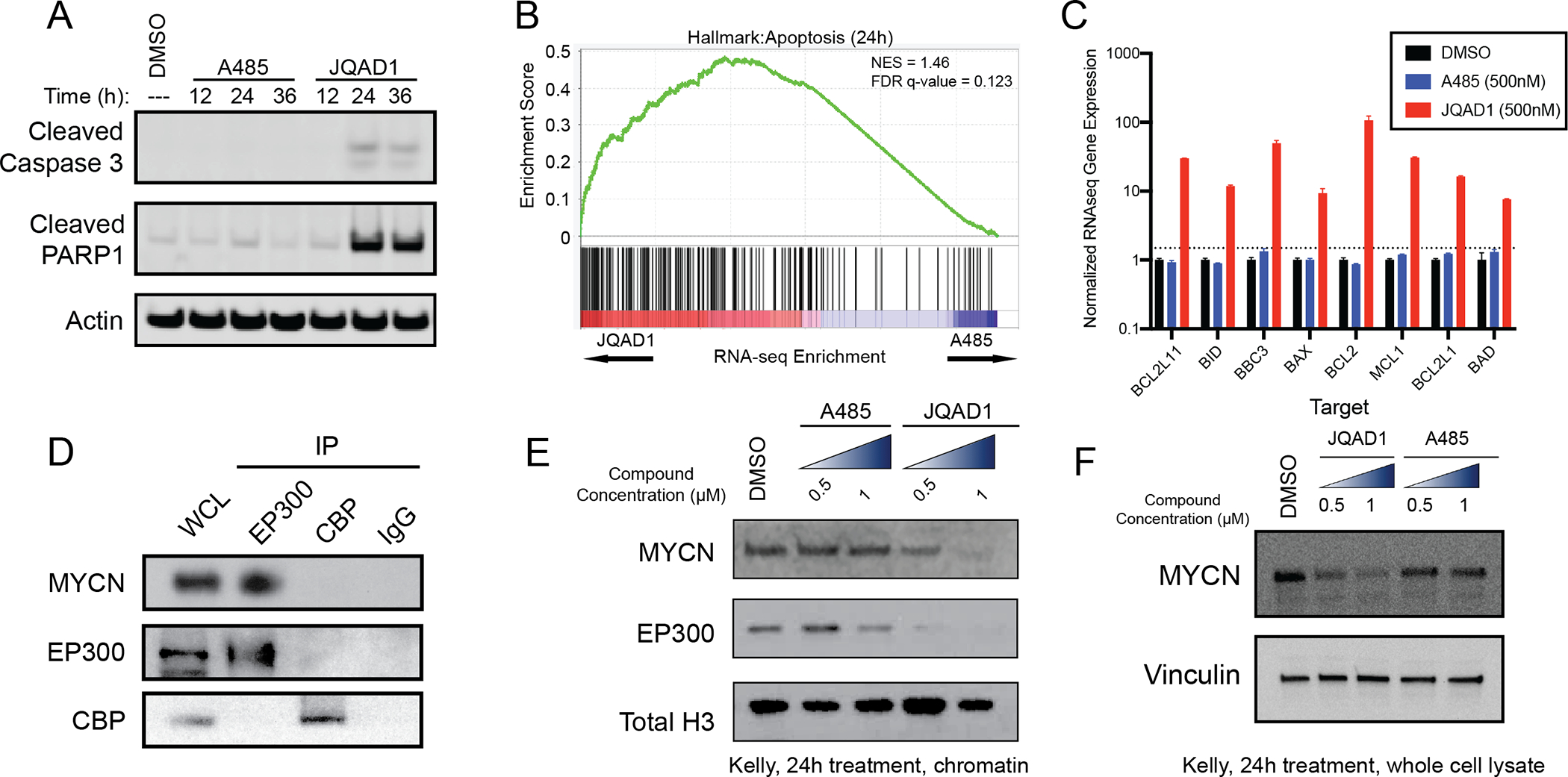
A. Kelly NB cells were treated with 1μM JQAD1, A485 or DMSO control for 12, 24, or 36h, prior to lysis and western blotting for the markers of apoptosis: cleaved caspase-3 and cleaved PARP1. Actin is demonstrated as a loading control. Data is representative of three independent treatments and analyses in Kelly and NGP cells.
B. Kelly cells were treated with 500nM JQAD1, A485 or DMSO control for 24h prior to ERCC-controlled spike in RNAseq. Gene set enrichment analysis of RNAseq results was performed with the MSigDB Hallmarks dataset. n=3 biological replicates and independent RNA extractions per treatment.
C. Normalized RNAseq gene expression of pro- and anti-apopotic mRNA transcripts from Kelly cells treated as in B. Log10 transcript expression is shown, normalized against DMSO and ERCC controls. n=3 biological replicates and independent RNA extractions per treatment. Bars represent S.E.M.
D. Nuclear lysates from Kelly cells were immunoprecipitated with anti-EP300, anti-CBP or IgG control antibodies. WCL=whole cell lysate. Data is representative of >3 independent co-immunoprecipitation/western blots.
E,F. Kelly cells were treated with DMSO control, A485 (0.5, 1 μM) or JQAD1 (0.5, 1 μM), followed by extraction of chromatin (E) or whole cell lysates (F) and western blotting. Total H3 is shown as a loading control. Data is representative of 3 independent biological replicates. See also Fig. S4.
