Fig. 2 |. Image-translation-assisted segmentation in 3D (ITAS3D): a two-step pipeline for annotation-free 3D segmentation of prostate glands.
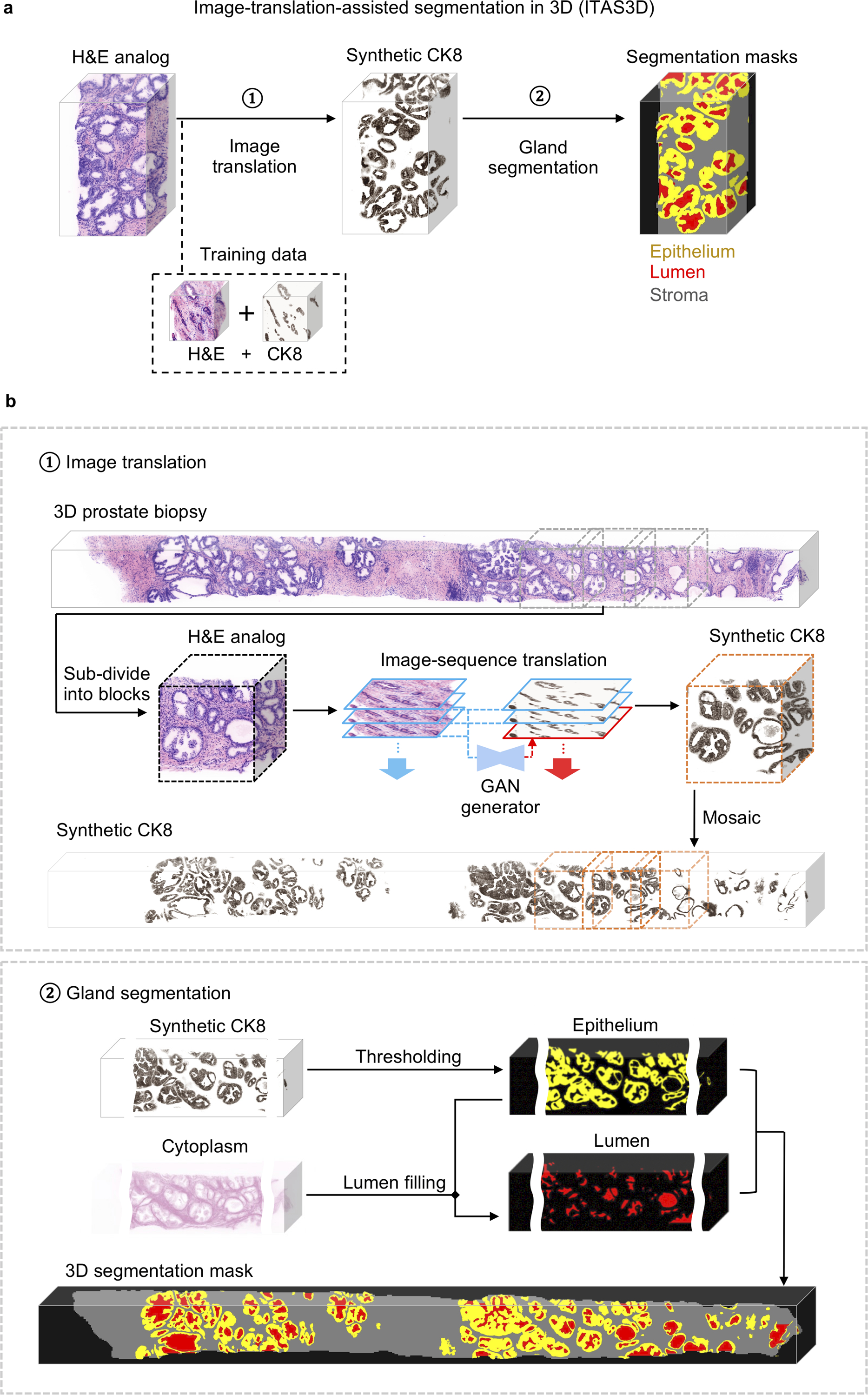
a, In step 1, a 3D microscopy dataset of a specimen, stained with a rapid and inexpensive fluorescent analog of H&E, is converted into a synthetic CK8 immunofluorescence (IF) dataset by using an image-sequence translation model that is trained with paired H&E-analog and real-CK8 IF datasets (tri-labeled tissues). The CK8 biomarker, which is utilized in standard-of-care genitourinary pathology practice, is ubiquitously expressed by the luminal epithelial cells of all prostate glands. In step 2, traditional computer-vision algorithms are applied to the synthetic-CK8 datasets for semantic segmentation of the gland epithelium, lumen, and surrounding stromal regions. b, In step 1, a 3D prostate biopsy is sub-divided into overlapping blocks that are each regarded as depth-wise sequences of 2D images. A GAN-trained generator performs image translation sequentially on each 2D level of an image block. The image translation at each level is based on the H&E-analog input at that level while leveraging the H&E-analog and CK8 images from two previous levels to enforce spatial continuity between levels (i.e., a “2.5D” translation method). The synthetic-CK8 image-block outputs are then mosaicked to generate a 3D CK8 dataset of the whole biopsy to assist with gland segmentation. In step 2, the epithelial cell layer (epithelium) is segmented from the synthetic-CK8 dataset with a thresholding-based algorithm. Gland lumen spaces are segmented by filling in the regions enclosed by the epithelia with refinements based on the cytoplasm channel (eosin fluorescence). See Supplementary Methods for details.
