Figure 1. Machine learning enables liver fat quantification and clinical and genetic analyses.
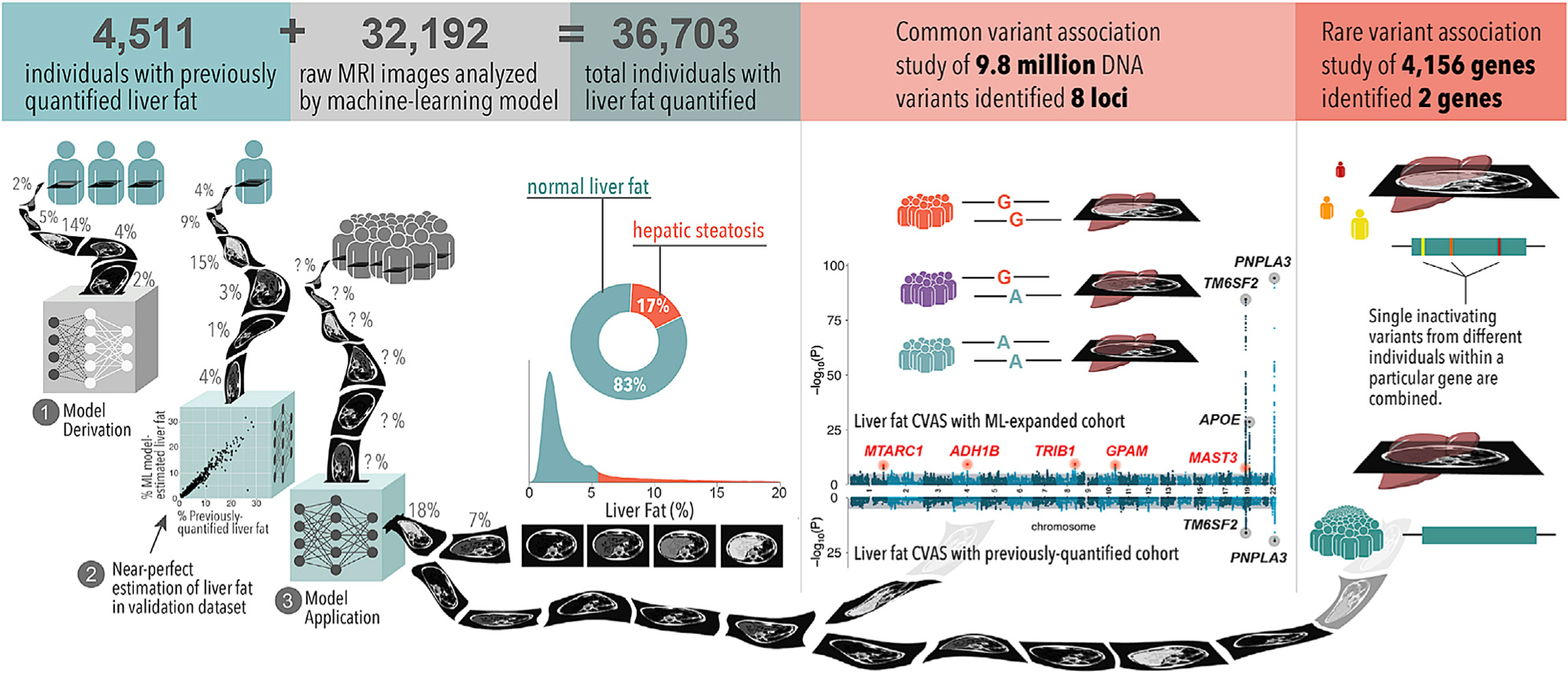
We developed a machine-learning model using a training set of 4,511 individuals with previously quantified liver fat from the UK Biobank. We applied this to estimate liver fat in an additional 32,192 individuals in the UK Biobank. Of the 36,703 total individuals with liver fat quantified, 17% met criteria for hepatic steatosis, defined as liver fat content greater than 5.5%. 1.6% of individuals had liver fat greater than 20% (not shown in the density plot). A common variant GWAS identified eight loci associated at genome-wide significance (p < 5.0 × 10−8), of which five are newly identified relative to previous studies (top Manhattan plot). None of these newly associated variants were identified in a common variant association study of those with liver fat quantified previously (bottom Manhattan plot). An RVAS identified inactivating variants in APOB and MTTP significantly (p < 1.2 × 10−5) associated with liver fat and steatosis.
