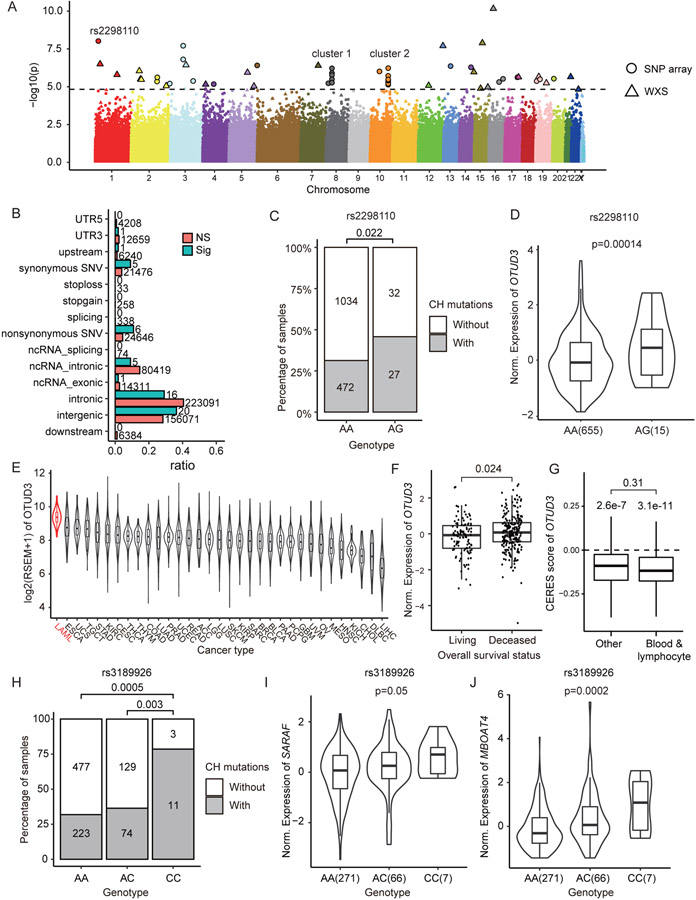
Figure 3. Genetic association of CH mutations.
(A) Manhattan plot showed 117 SNPs significantly associated with number of CH mutations. (B) Category of SNPs associated with CH mutations. Numbers on each bar denoted the number of SNPs in each category. Note that the total number of “Sig” SNPs were larger than 55, because a SNP might belong to multiple categories. (C) Number of samples with CH mutations correlated with genotype of rs2298110. Samples with heterozygous genotype AG tend to have more CH mutations. (D) In GTEx whole blood samples, expression of OTUD3 was positively correlated with heterozygous genotype AG of rs2298110. (E) Expression of OTUD3 among TCGA cancers. Acute myeloid leukemia (LAML) had the highest expression of OTUD3. (F) Higher expression of OTUD3 was correlated with poor overall survival status in TARGET leukemia data (31). (G) DepMap data suggested knockout OTUD3 broadly reduced the proliferation rate in various cell lines. While there were no significant CERES score differences between blood/lymphocyte cell lines and other cell lines (p=0.31), both kind of cell lines have CERES scores significantly lower than zero (p=3.1e-11 in blood/lymphocyte and 2.6e-7 in other cell lines respectively). (H) Number of samples with CH mutations correlated with genotype of rs3189926. Samples with homozygous genotype CC tend to have more CH mutations than genotype AA and AC. In GTEx whole blood data, samples with homozygous genotype CC at rs3189926 had higher expression of (I) SARAF and (J) MBOAT4 than other samples.