Extended Data Fig. 1. Schematic representation of LIBRA-seq experiments.
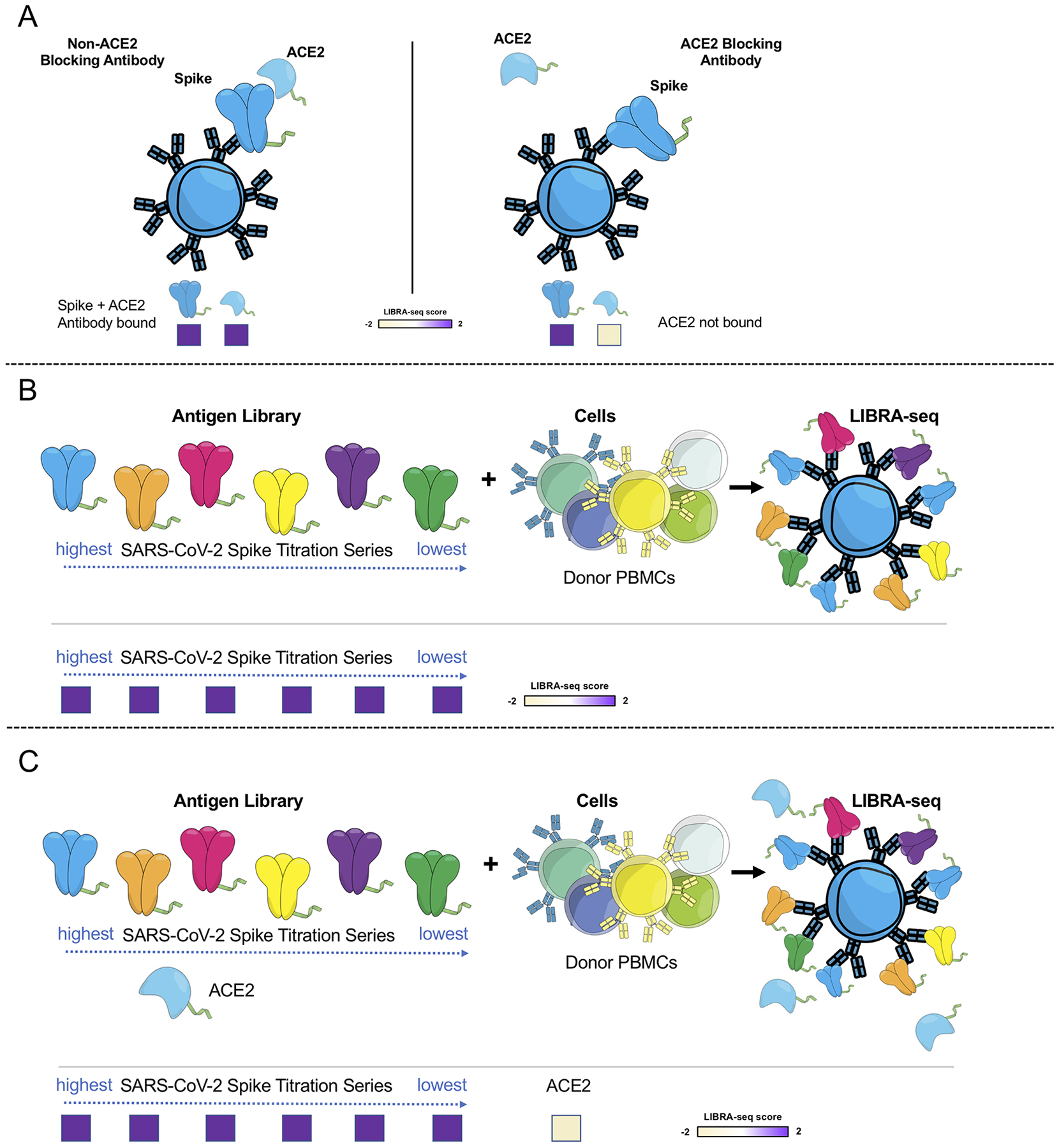
A. An antigen screening library of oligonucleotide-labeled antigens was generated. This library consisted of SARS-CoV-2 spike antigens and negative controls. Additionally, oligo-labeled ACE2 (the SARS-CoV-2 spike host cell receptor) was included. The antigen screening library was mixed with donor PBMCs. This approach allowed for assessment of B cell ligand blocking functionality from the sequencing experiment. B. An antigen screening library containing an antigen titration was generated, with a goal of identifying high affinity antibodies from LIBRA-seq. In this experiment, six different amounts of oligo-labeled SARS-CoV-2 S protein, each labeled with a different barcode, were included in a screening library. C. Schematic of LIBRA-seq with S titrations and ACE2 included for ligand blocking.
