Figure 5. QTL associated with UBB PC2.
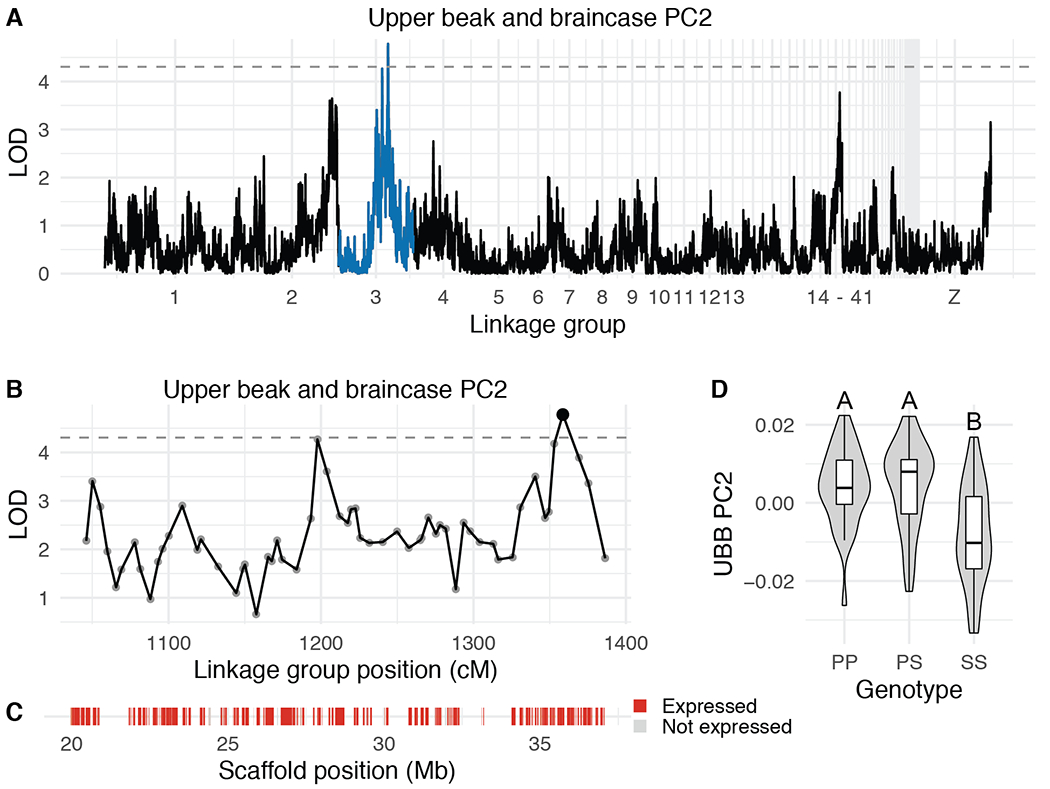
(A) Genome-wide QTL scan for UBB PC2. Dashed horizontal line indicates 5% genome-wide significance threshold and linkage groups with significant QTL peaks are highlighted in blue. (B) LOD support interval for UBB PC2 QTL scan. Dots indicate linkage map markers; the larger black dot highlights the peak marker that was used to estimate QTL effects. (C) Genes located within LOD support interval, color coded based on expression status in HH29 facial primordia. Expressed = transcript per kilobase million (TPM) ≥ 0.5; Not expressed = TPM < 0.5. (D) QTL effect plot for UBB PC2. Letters denote significance groups, p-values determined via Tukey test: PP vs. SS = 6.4e-04, PS vs. SS = 3.1e-05. P = allele from Pom founder, S = allele from Scan founder.
