Figure 4.
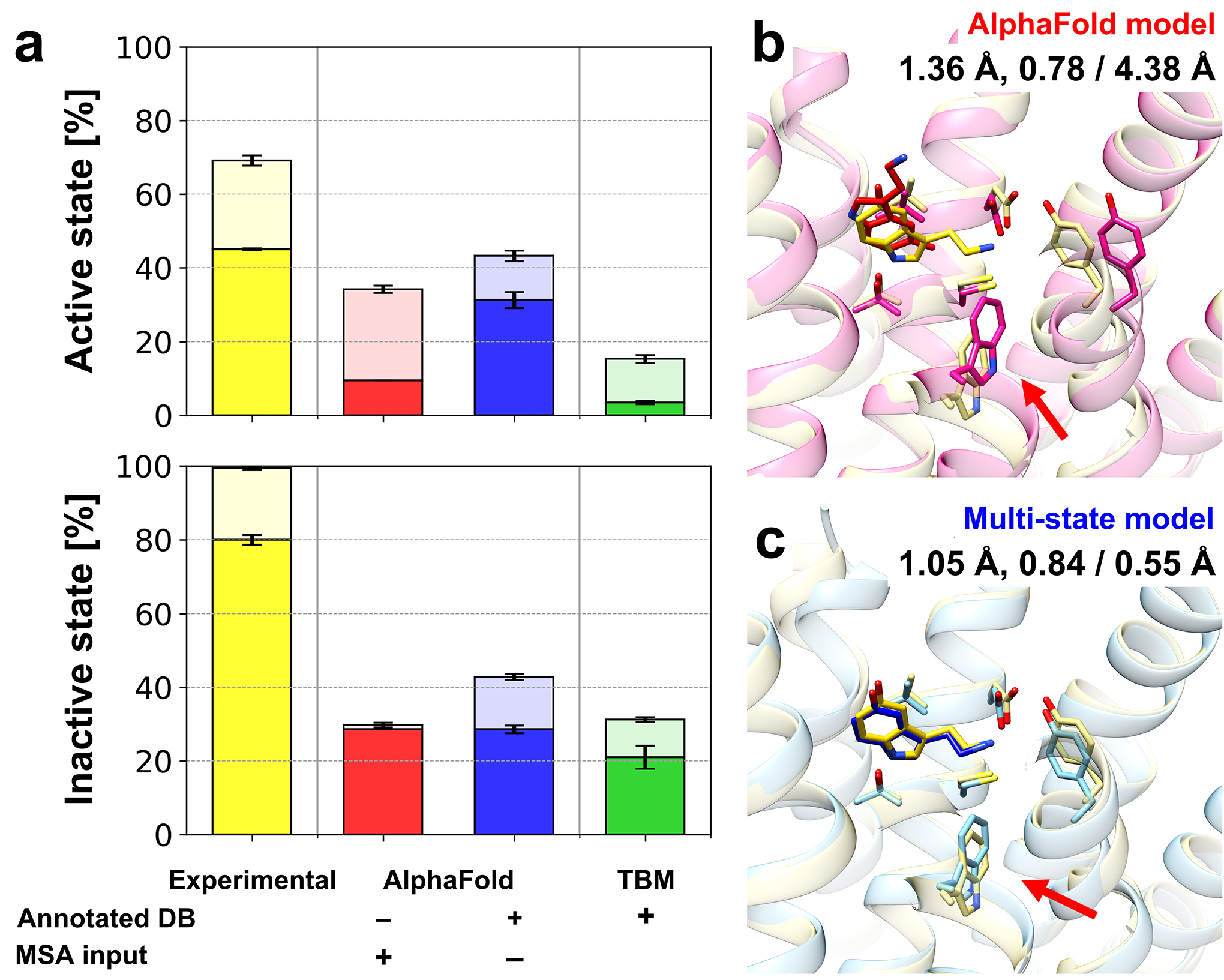
Protein-ligand docking on predicted GPCR models. (a) Protein-ligand docking success ratios for GPCR model structures using various modeling protocols. Docking simulations were performed five times independently for each structure using AutoDock Vina. Success ratios and their standard errors are shown as bar charts for top 1 (opaque) and top 3 (transparent) predictions and overlaid error bars, respectively. An example of protein-ligand docking using different models is presented: (b) the original AlphaFold model and (c) multi-state model. An endogenous agonist ligand (serotonin, 5-hydroxytryptamine, SRO from PDB ID 7E2Y) was docked to protein structure models for human 5-hydroxytrypamine receptor 1A (UniProt ID P08908, 5HT1A_HUMAN). Protein structures are shown in transparent cartoon representations, while docked ligand conformations are depicted as sticks. The experimental protein-ligand complex structure is shown in yellow. Key structural differences between the two protein models that contributed to the different docking performance are indicated by red arrows. The binding site heavy-atom RMSD, the lDDT for the transmembrane region, and the resulting docking accuracy in terms of ligand heavy-atom RMSD are shown under the model names.
