Figure 4.
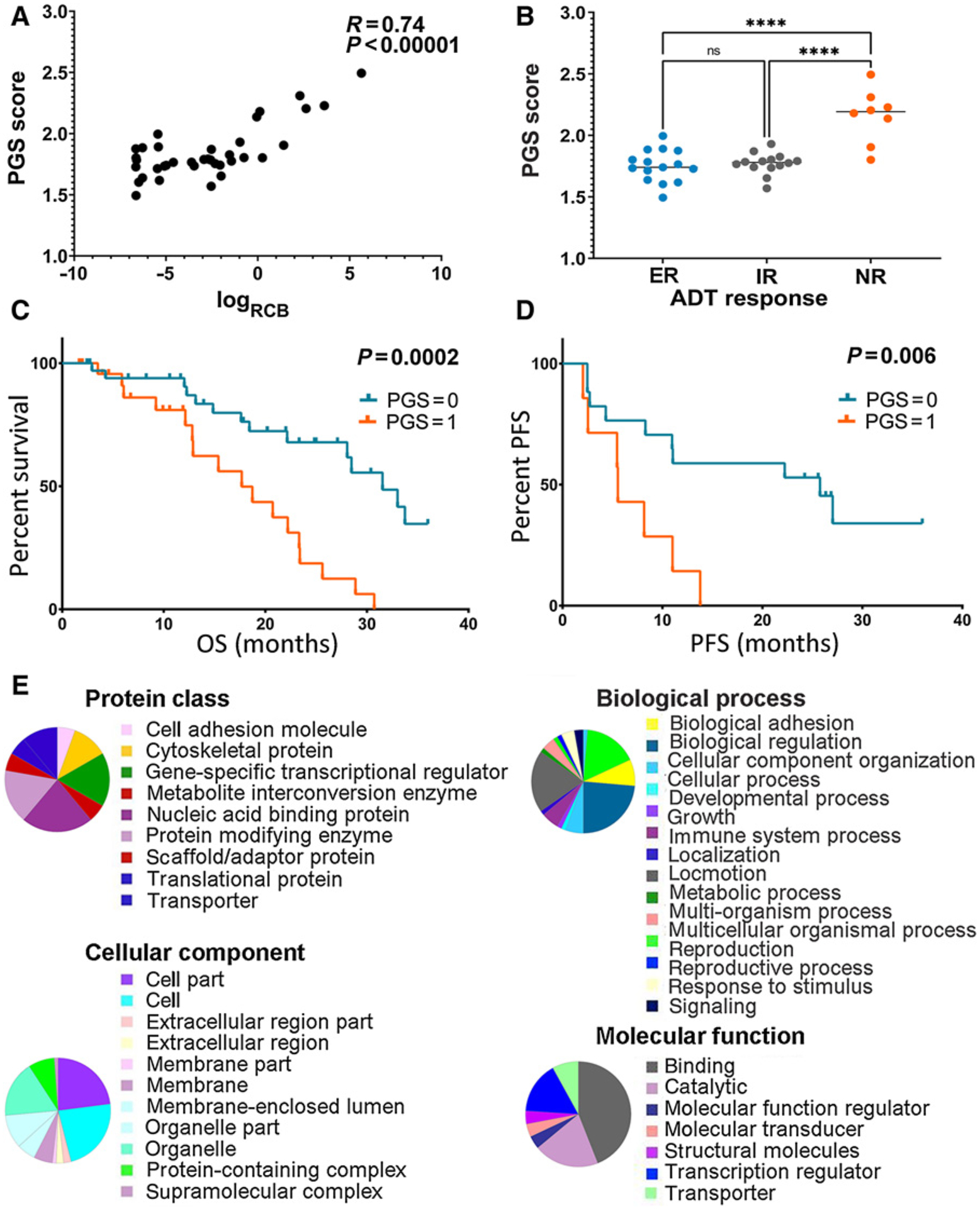
PGS predicted ADT response or clinical outcomes in independent hormone-naïve or CRPC clinical cohorts. A, Single-sample gene set enrichment score of PGS is proportionally related to the residual tumor volumes [RCB, r(35) = 0.74, P < 0.00001] in primary hormone-naïve prostate cancer treated with preoperative ADT (Sowalsky clinical cohort, n = 37). B, In the Sowalsky cohort, the PGS gene set enrichment score is significantly higher in nonresponders (NR) compared with exceptional responders (ER; P < 0.0001) and incomplete responders (IR; P < 0.0001). C and D, Kaplan–Meier estimate of OS or PFS segregated according to the PGS score in SU2C (A, n = 60) and Alumkal (B, n = 25) clinical cohorts. In both cohorts, patients with PGS = 1 had worse outcome than those with PGS = 0 (P = 0.0002 and 0.006, respectively). E, Panther Protein Classification of the PGS genes revealed that the genes were involved in various biological processes.
