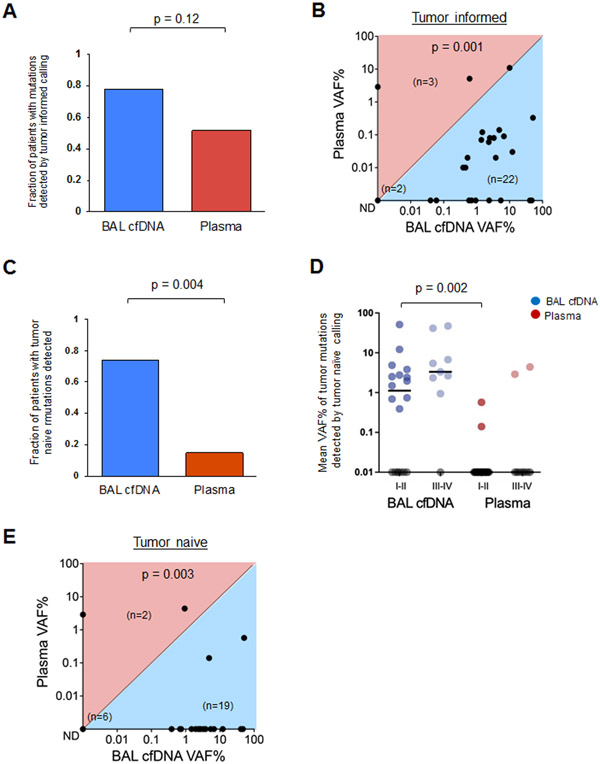
Figure 2. Bronchoalveolar lavage identifies more tumor mutations than plasma.
(A) Fraction of patients with tumor-derived mutations identified in BAL cfDNA (blue) compared to plasma cfDNA (red) using the tumor informed approach for 27 patients (Supplementary Table 5).
(B) Mean Variant Allele Frequency (mean VAF%) in BAL and plasma cfDNA using the tumor informed approach to identify tumor-derived variants. Each dot represents one patient. Numbers of patients in each half of the plot are shown in parentheses within the plot along with the number of samples with no tumor derived mutations detected (ND).
(C) Driver Mutations identified in BAL cfDNA (blue) compared to plasma cfDNA (red) using the tumor naive calling approach for 27 patients (Supplemental Table 7).
(D) Mean VAF% of mutant DNA detected in BAL cfDNA (blue) or plasma cfDNA (red) by stage using the tumor naïve calling strategy (p = 0.002). Each dot represents one patient and samples with no mutations detected (ND) are shown on the x-axis in gray.
(E) Mean VAF% in BAL and plasma cfDNA using the tumor naïve approach. Each dot represents one patient. Numbers of patients in each half of the plot are shown in parentheses within the plot along with the number of samples with no tumor derived mutations detected (ND).