Fig 4: Discovery of DNA repair and replication components enriched in non-pCR TNBC tumors.
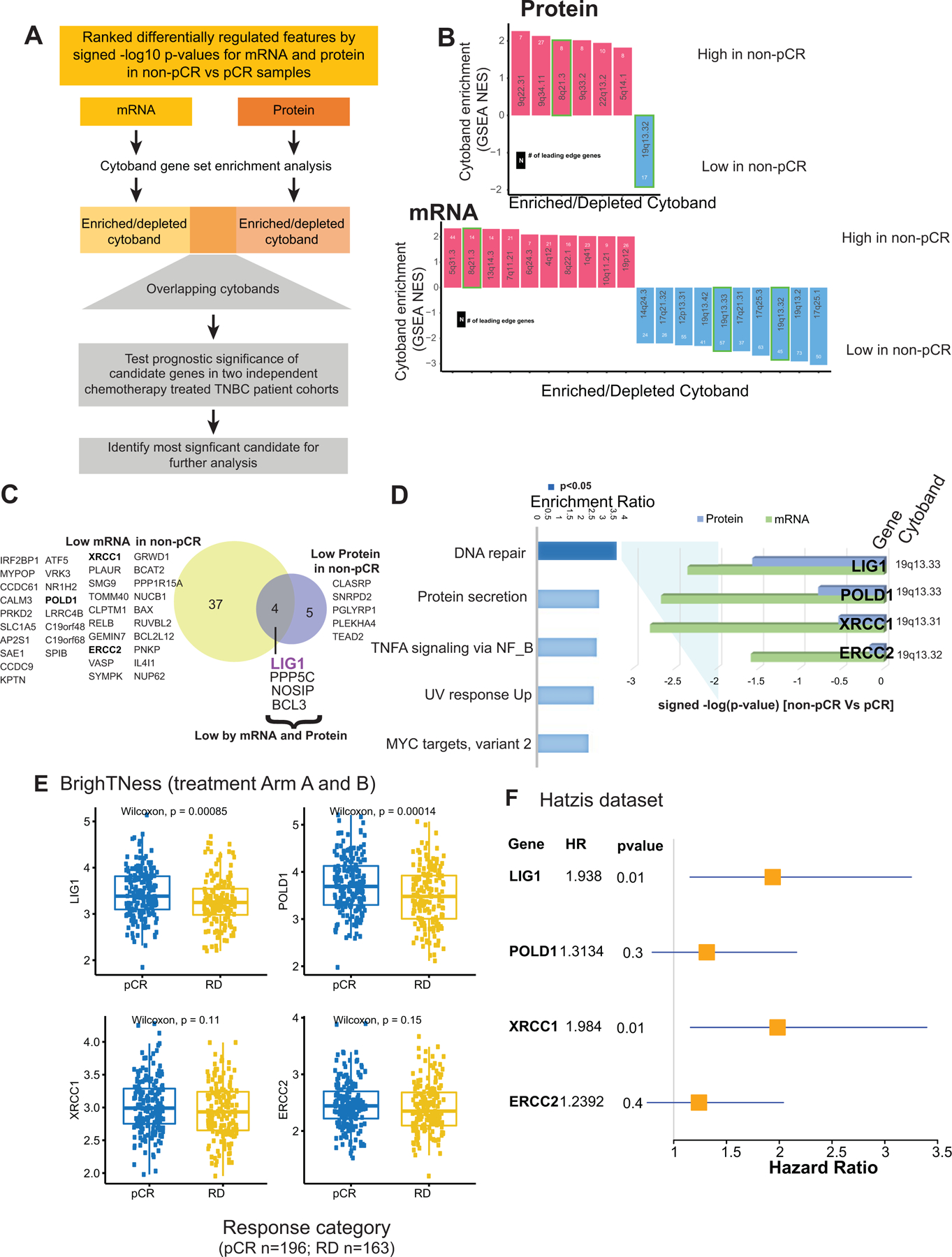
A) p=Cytobands enriched in genes differentially expressed between non-pCR and pCR for both mRNA and protein. To identify up or downregulated features over-represented in certain cytobands within the chromosome, GSEA was used to identify regions from chromosomal location databases enriched with differential genes (GSEA input was ranked expression list (signed -log10 pvalue) from Wilcoxon rank sum tests). Overrepresented cytobands that were either enriched or depleted using differentially expressed mRNA and protein are indicated in B, and the overlapping sets were used for further analysis. Genes downregulated in non-pCR samples correspond to cytoband 19q13.31–33 is indicated in C.
B) Plot showing significantly enriched or depleted cytobands obtained by running differential mRNA and protein ranked lists through GSEA.
C) Venn-diagram showing differential (non-pCR vs. pCR) mRNA and proteins located on cytoband 19q13.3.
D) Over-representation analysis (ORA) shows that differential 19q13.31–33 genes are enriched with Hallmark DNA repair pathway genes. Downregulation of these DNA repair genes at mRNA and protein level in non-pCR cases is shown in the bar chart on the right as signed -log10 p-values from Wilcoxon rank sum tests.
E) Boxplot showing comparing RNA expression of DNA-repair genes located on 19q13.31–33 in the previously published BrighTNess clinical trial (Treatment Arm A and B), where patients were treated with carboplatin and paclitaxel. The Wilcoxon rank sum test was used to compare residual disease (RD) cases to pCR cases.
F) Forest plot showing hazard ratios (HR) and p-values for metastasis free survival associated with LIG1, POLD1, XRCC1 and ERCC2. HR is based on categorizing samples using median expression cutoff for each gene in the Hatzis dataset.
