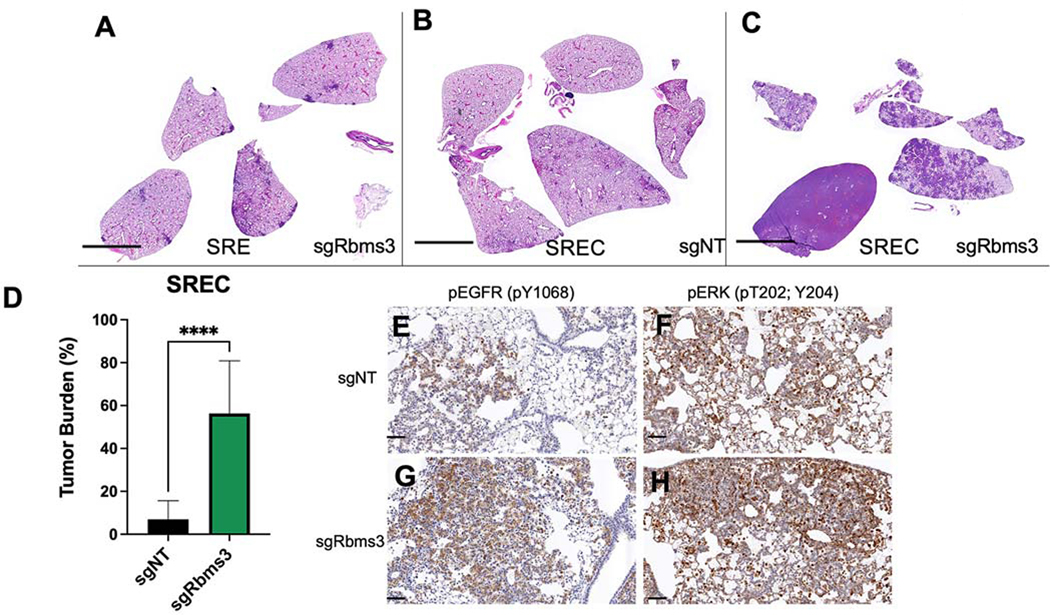
Figure 5. Rbms3 loss cooperates with EGFRL858R to accelerate malignant lung adenocarcinoma.
A-C: Images of harvested mouse lung sections following necropsy analyses stained with hematoxylin and eosin (H&E) 11 weeks post initiation with 1 × 105 pfu lenti-CRE, followed by continuous administration of doxycycline chow to induce EGFRL858R expression. CRISPR/CAS9-mediated genome editing was used in panels B and C to edit Rbms3 in vivo. Genotype of each experimental group was: A: sgRbms3-CRE virus in SPC::CRE-ERT2/+; Rosa26CAGs-LSL-rTTa3; EGFRL858R (SRE) mice. B: sgNT-CRE virus in or SPC::CRE-ERT2/+; Rosa26CAGs-LSL-rTTa3; EGFRL858R; H11LSL-CAS9/+ (SREC) mice. C: sgRbms3-CRE virus in SREC mice. Black bar in bottom left of each panel represents a 1000-micron scale bar. D: Quantification of tumor burden from genotypes in panel B compared to panel C. N=5 mice per group. The mean is graphed, and error bars represent SEM. Statistical analysis was conducted using a paired T test; **** = p < 0.0001. E: Representative images of immunohistochemistry on FFPE lung tissue sections from SREC mice initiated with either sgNT- or sgRbms3-CRE and stained with pEGFR (pY1068) or pERK (pT202; Y204) shown at 20x magnification. Scale bar shown in black at the bottom left corner of each image represent 50 microns.