Figure 3. NF-YA contributes to genomic specificity of the Menin-UTX molecular switch on chromatin.
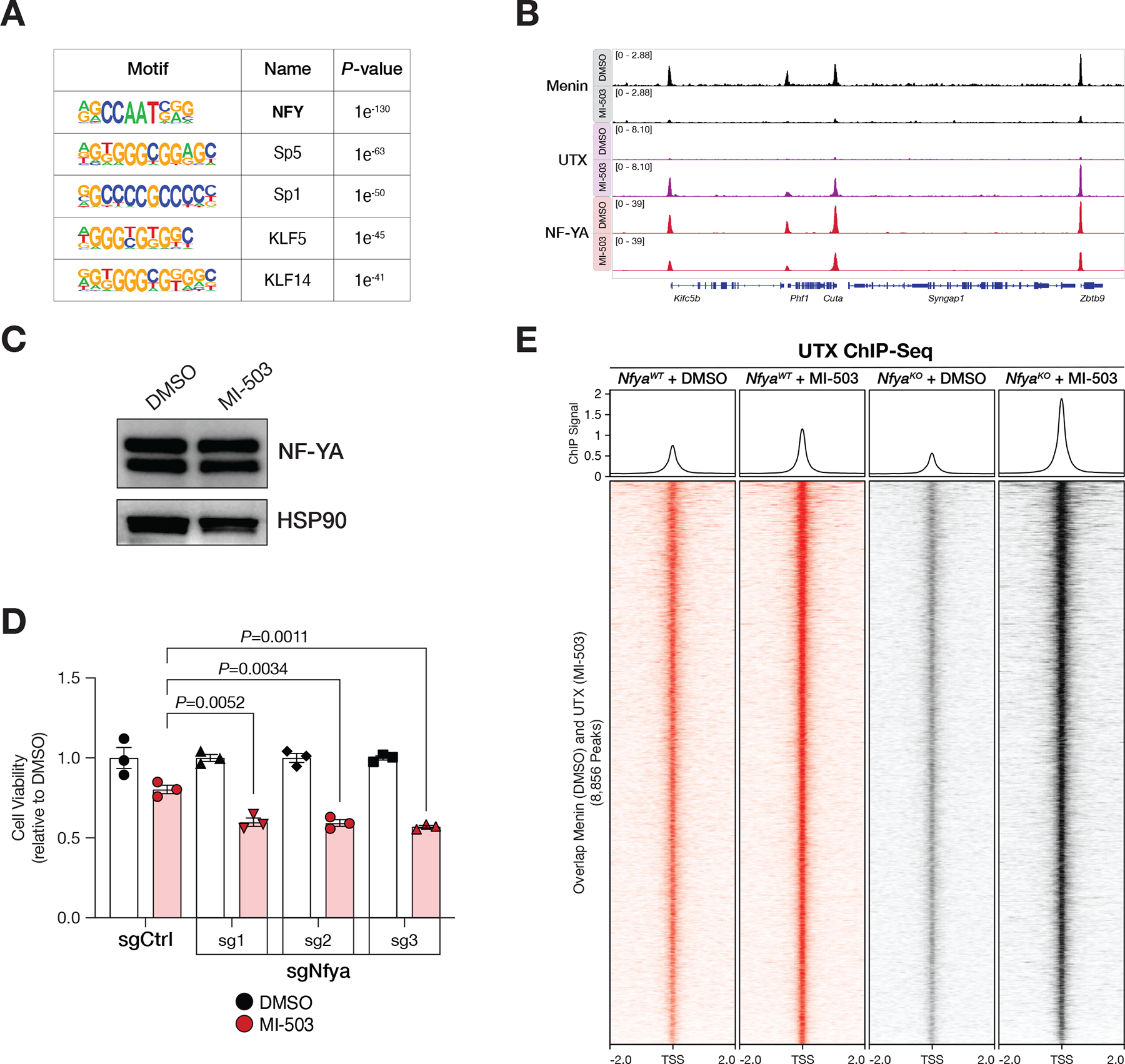
(A) HOMER de novo motif analysis of overlapping ChIP-Seq peaks between Menin (in DMSO) and UTX (in MI-503) in mouse MLL-AF9 leukemia cells. (B) Genome browser representation of ChIP-Seq normalized reads (average RPKM) for representative loci bound by Menin (black), UTX (purple), and NF-YA (red) in cells treated with vehicle (DMSO) or Menin-MLL inhibitor (MI-503) for 96 hours. (C) Immunoblot analysis of NF-YA and HSP90 proteins (loading control) upon Menin-MLL inhibitor (MI-503) treatment of mouse MLL-AF9 leukemia cells for 96 hours. (D) Viability assay from cells treated with vehicle (DMSO, black) or Menin-MLL inhibitor (MI-503, red) for 96 hours (mean±SEM, n=3 infection replicates, P-value calculated by Student’s t-test). sgCtrl, control sgRNA targeting a non-genic region on chromosome 8. (E) Heatmaps displaying UTX ChIP-Seq signal mapping to a 4-kb window around TSS in NfyaWT (red) or NfyaKO (black) mouse MLL-AF9 leukemia cells treated with vehicle (DMSO) or Menin-MLL inhibitor (MI-503) for 96 hours. Metaplot represents the average UTX ChIP-Seq signal at promoters.
