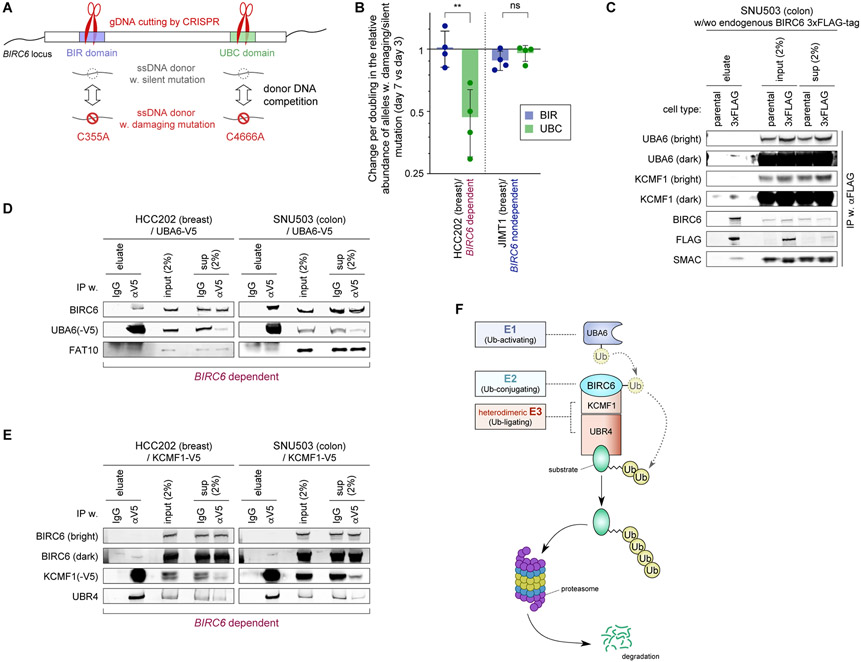
Figure 3. Biochemical demonstration of the BIRC6 complex assembly.
A. Competition assay to evaluate the essentiality of each of the two functional domains of BIRC6 using a strategy to repair a CRISPR-mediated cleavage of the genomic locus corresponding to each of these domains (BIR and UBC) via homologous recombination. We show the two different donor DNAs that were introduced, one harboring a damaging mutation and the other containing a silent mutation. This assay scores the relative abundance of alleles with damaging versus silent mutations.
B. Relative abundance of the damaging versus silent mutations in each of the two functional domains of BIRC6. Plotted are the change in the ratio of damaging over silent mutations at day seven after the transduction of the Cas9/crRNA ribonucleoprotein complex relative to the corresponding ratio at day three, normalized against the doubling time of the cells. Values = means ± SD (n = 4). ns p ≥ 0.05, **p <0.01.
C-E. Protein-protein interactions between the components of the BIRC6 module. In C, endogenously expressed BIRC6 was immunoprecipitated from the lysate of SNU503 cells that were engineered to have the 3xFLAG-tag-encoding sequence inserted at the N-terminus of BIRC6-encoding sequence. In D and E, exogenously-expressed, V5-tagged UBA6 (D) and V5-tagged KCMF1 (E) were immunoprecipitated from the lysates of HCC202 and SNU503 cells. In all these cases, eluate, crude (input) and cleared (sup) lysates were analyzed by immunoblotting.
F. The BIRC6 module is composed of an E1 enzyme (UBA6), an E2 enzyme (BIRC6), and two E3 enzymes that have been shown to work cooperatively (KCMF1 and UBR4).
All the experiments were performed twice, except for B, which shows the summary of four independent experiments.