Figure 2. Collaborative regulation of the BENC module underlies convergent differentiation induced by SWI/SNF or PU.1 inhibition in AML.
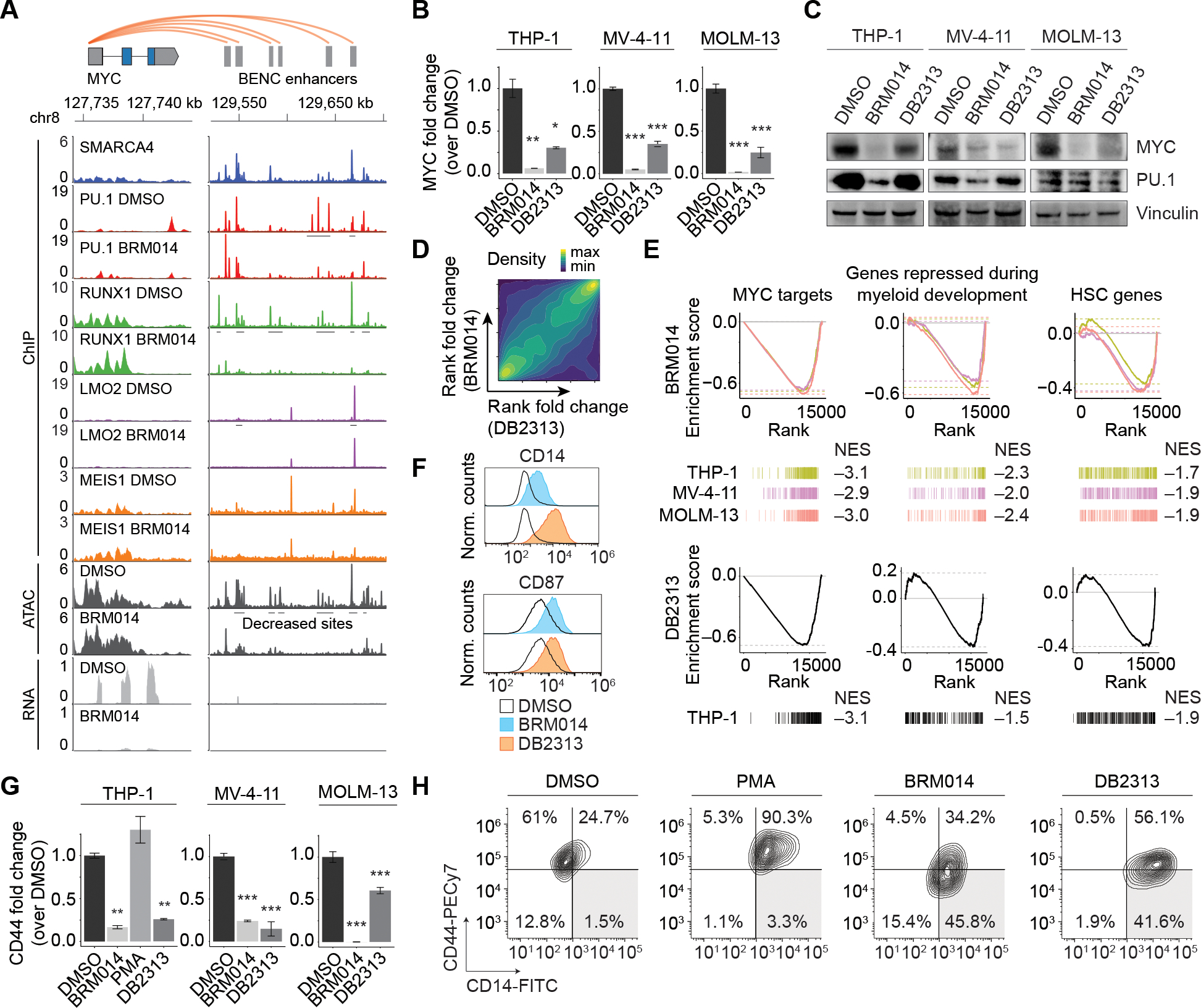
(A) Overlay of SMARCA4, PU.1, RUNX1, LMO2, and MEIS1 occupancy with ATAC-seq and RNA-seq data at MYC and BENC in THP-1 cells. Other cell lines presented in Figure S3A–B. Expression of MYC and PU.1 in AML cells lines treated with BRM014 or DB2313 by (B) RNA-seq (THP-1) or RT-qPCR (MV-4–11 and MOLM-13) at 72 h, N=3, and (C) Western blot at 24 h of treatment. (D) Genome-wide correlation of BRM014- and DB2313-induced transcriptional changes in THP-1 cells based on RNA-seq. (E) Gene sets downregulated upon BRM014 and DB2313 treatment. Normalized enrichment scores (NES) are indicated. (F) Cell surface expression of myeloid differentiation markers CD14 and CD87 in cells treated with DMSO, BRM014, or DB2313. (G) Expression of CD44 in cells treated with DMSO, BRM014, DB2313, or PMA measured by RNA-seq (THP-1) or RT-qPCR (MV-4–11 and MOLM-13) at 72 h, N = 3. (H) Expression of CD14 and CD44 in cells treated with DMSO, BRM014, DB2313, or PMA measured by flow cytometry. Error bars: mean ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001.
