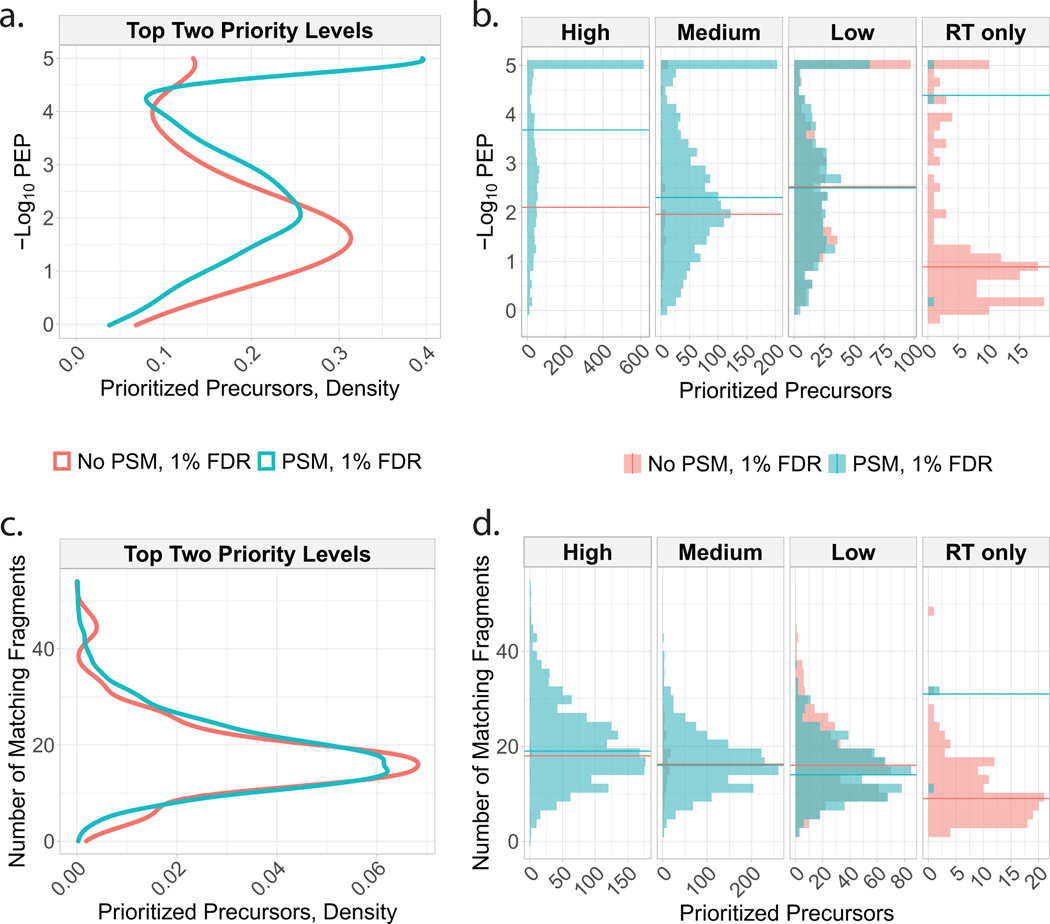
Extended Data Fig. 3 |. Properties of peptides successfully identified in pSCoPE runs.
The precursors from the inclusion list were split into those that resulted in confident PSMs and those that did not, and the properties of each set analyzed based on the shotgun runs used for making the inclusion lists. (a) Confidence of identification (quantified by the posterior error probability; PEP) and number of matching peptide fragments for successful and unsuccessful precursors. The data are shown for all prioritized peptides across all priority tiers. (b) The data from panel a are shown faceted by priority tier. All data shown are from the consistency experiments from Fig. 2c. In previous analyses conducted during a period of suboptimal instrument performance, the number of matching fragments was shown to effectively distinguish between the peptides which were identified at 1\% FDR and those that were not identified, which was reported in version 1 of our preprint. This trend is not observed in the current dataset, which was acquired by the same instrument but with more efficient ion isolation by its quadrupole, (c) and (d).